[English] 日本語
 Yorodumi
Yorodumi- EMDB-43655: Cryo-EM structure of human core Rab3GAP1/2 complex, local refinement -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human core Rab3GAP1/2 complex, local refinement | |||||||||
 Map data Map data | Local refinement from CryoSPARC, z-flipped | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / GAP / GEF / Rab3GAP / Lipid droplet / ER / Rab regulator / membrane trafficking / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of glutamate neurotransmitter secretion in response to membrane depolarization / positive regulation of endoplasmic reticulum tubular network organization / establishment of protein localization to endoplasmic reticulum membrane / regulation of calcium ion-dependent exocytosis of neurotransmitter / synaptic signaling / endoplasmic reticulum tubular network / positive regulation of protein lipidation / positive regulation of autophagosome assembly / lipid droplet organization / RAB GEFs exchange GTP for GDP on RABs ...positive regulation of glutamate neurotransmitter secretion in response to membrane depolarization / positive regulation of endoplasmic reticulum tubular network organization / establishment of protein localization to endoplasmic reticulum membrane / regulation of calcium ion-dependent exocytosis of neurotransmitter / synaptic signaling / endoplasmic reticulum tubular network / positive regulation of protein lipidation / positive regulation of autophagosome assembly / lipid droplet organization / RAB GEFs exchange GTP for GDP on RABs / camera-type eye development / regulation of short-term neuronal synaptic plasticity / COPI-independent Golgi-to-ER retrograde traffic / face morphogenesis / regulation of GTPase activity / hypothalamus development / enzyme regulator activity / lipid droplet / GTPase activator activity / autophagosome / positive regulation of GTPase activity / enzyme activator activity / guanyl-nucleotide exchange factor activity / excitatory postsynaptic potential / intracellular protein transport / macroautophagy / brain development / small GTPase binding / presynaptic membrane / postsynapse / endoplasmic reticulum membrane / Golgi apparatus / protein-containing complex / extracellular exosome / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
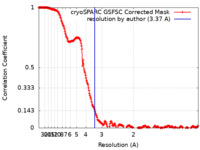
| Method | single particle reconstruction / cryo EM / Resolution: 3.37 Å | |||||||||
 Authors Authors | Nguyen KM / Yip CK | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2025 Journal: Nat Commun / Year: 2025Title: Biochemical and structural characterization of Rab3GAP reveals insights into Rab18 nucleotide exchange activity. Authors: Gage M J Fairlie / Kha M Nguyen / Sung-Eun Nam / Alexandria L Shaw / Matthew A H Parson / Hannah R Shariati / Xinyin Wang / Meredith L Jenkins / Michael Gong / John E Burke / Calvin K Yip /  Abstract: The heterodimeric Rab3GAP complex is a guanine nucleotide exchange factor (GEF) for the Rab18 GTPase that regulates lipid droplet metabolism, ER-to-Golgi trafficking, secretion, and autophagy. Why ...The heterodimeric Rab3GAP complex is a guanine nucleotide exchange factor (GEF) for the Rab18 GTPase that regulates lipid droplet metabolism, ER-to-Golgi trafficking, secretion, and autophagy. Why both subunits of Rab3GAP are required for Rab18 GEF activity and the molecular basis of how Rab3GAP engages and activates its cognate substrate are unknown. Here we show that human Rab3GAP is conformationally flexible and potentially autoinhibited by the C-terminal domain of its Rab3GAP2 subunit. Our high-resolution structure of the catalytic core of Rab3GAP, determined by cryo-EM, shows that the Rab3GAP2 N-terminal domain binds Rab3GAP1 via an extensive interface. AlphaFold3 modelling analysis together with targeted mutagenesis and in vitro activity assay reveal that Rab3GAP likely engages its substrate Rab18 through an interface away from the switch and interswitch regions. Lastly, we find that three Warburg Micro Syndrome-associated missense mutations do not affect the overall architecture of Rab3GAP but instead likely interfere with substrate binding. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43655.map.gz emd_43655.map.gz | 495.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43655-v30.xml emd-43655-v30.xml emd-43655.xml emd-43655.xml | 23.1 KB 23.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43655_fsc.xml emd_43655_fsc.xml | 21.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_43655.png emd_43655.png | 45.3 KB | ||
| Filedesc metadata |  emd-43655.cif.gz emd-43655.cif.gz | 7.3 KB | ||
| Others |  emd_43655_additional_1.map.gz emd_43655_additional_1.map.gz emd_43655_half_map_1.map.gz emd_43655_half_map_1.map.gz emd_43655_half_map_2.map.gz emd_43655_half_map_2.map.gz | 912.9 MB 929.1 MB 929.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43655 http://ftp.pdbj.org/pub/emdb/structures/EMD-43655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43655 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43655 | HTTPS FTP |
-Validation report
| Summary document |  emd_43655_validation.pdf.gz emd_43655_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43655_full_validation.pdf.gz emd_43655_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_43655_validation.xml.gz emd_43655_validation.xml.gz | 31.1 KB | Display | |
| Data in CIF |  emd_43655_validation.cif.gz emd_43655_validation.cif.gz | 40.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43655 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43655 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43655 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43655 | HTTPS FTP |
-Related structure data
| Related structure data |  8vybMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43655.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43655.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local refinement from CryoSPARC, z-flipped | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.59 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Auto-sharpened map from Phenix
| File | emd_43655_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Auto-sharpened map from Phenix | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A, z-flipped
| File | emd_43655_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A, z-flipped | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map B, z-flipped
| File | emd_43655_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B, z-flipped | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Core Rab3GAP1/2 complex
| Entire | Name: Core Rab3GAP1/2 complex |
|---|---|
| Components |
|
-Supramolecule #1: Core Rab3GAP1/2 complex
| Supramolecule | Name: Core Rab3GAP1/2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Rab3GAP1
| Supramolecule | Name: Rab3GAP1 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Rab3GAP2
| Supramolecule | Name: Rab3GAP2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Isoform 2 of Rab3 GTPase-activating protein catalytic subunit
| Macromolecule | Name: Isoform 2 of Rab3 GTPase-activating protein catalytic subunit type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 116.747188 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MWSHPQFEKG GGSGGGSGGG SWSHPQFEKL EVLFQGPRSE ARGIQRPTST MAADSEPESE VFEITDFTTA SEWERFISKV EEVLNDWKL IGNSLGKPLE KGIFTSGTWE EKSDEISFAD FKFSVTHHYL VQESTDKEGK DELLEDVVPQ SMQDLLGMNN D FPPRAHCL ...String: MWSHPQFEKG GGSGGGSGGG SWSHPQFEKL EVLFQGPRSE ARGIQRPTST MAADSEPESE VFEITDFTTA SEWERFISKV EEVLNDWKL IGNSLGKPLE KGIFTSGTWE EKSDEISFAD FKFSVTHHYL VQESTDKEGK DELLEDVVPQ SMQDLLGMNN D FPPRAHCL VRWYGLREFV VIAPAAHSDA VLSESKCNLL LSSVSIALGN TGCQVPLFVQ IHHKWRRMYV GECQGPGVRT DF EMVHLRK VPNQYTHLSG LLDIFKSKIG CPLTPLPPVS IAIRFTYVLQ DWQQYFWPQQ PPDIDALVGG EVGGLEFGKL PFG ACEDPI SELHLATTWP HLTEGIIVDN DVYSDLDPIQ APHWSVRVRK AENPQCLLGD FVTEFFKICR RKESTDEILG RSAF EEEGK ETADITHALS KLTEPASVPI HKLSVSNMVH TAKKKIRKHR GVEESPLNND VLNTILLFLF PDAVSEKPLD GTTST DNNN PPSESEDYNL YNQFKSAPSD SLTYKLALCL CMINFYHGGL KGVAHLWQEF VLEMRFRWEN NFLIPGLASG PPDLRC CLL HQKLQMLNCC IERKKARDEG KKTSASDVTN IYPGDAGKAG DQLVPDNLKE TDKEKGEVGK SWDSWSDSEE EFFECLS DT EELKGNGQES GKKGGPKEMA NLRPEGRLYQ HGKLTLLHNG EPLYIPVTQE PAPMTEDLLE EQSEVLAKLG TSAEGAHL R ARMQSACLLS DMESFKAANP GCSLEDFVRW YSPRDYIEEE VIDEKGNVVL KGELSARMKI PSNMWVEAWE TAKPIPARR QRRLFDDTRE AEKVLHYLAI QKPADLARHL LPCVIHAAVL KVKEEESLEN ISSVKKIIKQ IISHSSKVLH FPNPEDKKLE EIIHQITNV EALIARARSL KAKFGTEKCE QEEEKEDLER FVSCLLEQPE VLVTGAGRGH AGRIIHKLFV NAQRLTESSD E AAAMTPPE EELKRMGSPE ERRQNSVSDF PPPAGREFIL RTTVPRPAPY SKALPQRMYS VLTKEDFRLA GAFSSDTSFF UniProtKB: Rab3 GTPase-activating protein catalytic subunit |
-Macromolecule #2: Rab3 GTPase-activating protein non-catalytic subunit
| Macromolecule | Name: Rab3 GTPase-activating protein non-catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 65.121422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MACSIVQFCY FQDLQAARDF LFPHLREEIL SGALRRDPSK STDWEDDGWG AWEENEPQEP EEEGNTCKTQ KTSWLQDCVL SLSPTNDLM VIAREQKAVF LVPKWKYSDK GKEEMQFAVG WSGSLNVEEG ECVTSALCIP LASQKRSSTG RPDWTCIVVG F TSGYVRFY ...String: MACSIVQFCY FQDLQAARDF LFPHLREEIL SGALRRDPSK STDWEDDGWG AWEENEPQEP EEEGNTCKTQ KTSWLQDCVL SLSPTNDLM VIAREQKAVF LVPKWKYSDK GKEEMQFAVG WSGSLNVEEG ECVTSALCIP LASQKRSSTG RPDWTCIVVG F TSGYVRFY TENGVLLLAQ LLNEDPVLQL KCRTYEIPRH PGVTEQNEEL SILYPAAIVT IDGFSLFQSL RACRNQVAKA AA SGNENIQ PPPLAYKKWG LQDIDTIIDH ASVGIMTLSP FDQMKTASNI GGFNAAIKNS PPAMSQYITV GSNPFTGFFY ALE GSTQPL LSHVALAVAS KLTSALFNAA SGWLGWKSKH EEEAVQKQKP KVEPATPLAV RFGLPDSRRH GESICLSPCN TLAA VTDDF GRVILLDVAR GIAIRMWKGY RDAQIGWIQT VEDLHERVPE KADFSPFGNS QGPSRVAQFL VIYAPRRGIL EVWST QQGP RVGAFNVGKH CRLLYPGYKI MGLNNVTSQS WQPQTYQICL VDPVSGSVKT VNVPFHLALS DKKLREQKLE LGGSGG RQL DYKDHDGDYK DHDIDYKDDD DK UniProtKB: Rab3 GTPase-activating protein non-catalytic subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat-2/1 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 25 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 2s blot time, 5s blot force. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 215000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8vyb: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)