[English] 日本語
 Yorodumi
Yorodumi- EMDB-43227: DNA origami colloid for self-assembly of tubules: (10,0) monomer -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | DNA origami colloid for self-assembly of tubules: (10,0) monomer | |||||||||
 Map data Map data | Primary map of the (10,0) monomer. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA origami / synthetic construct / self-assembly / DNA | |||||||||
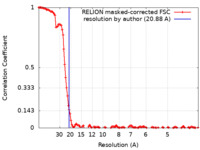
| Method | single particle reconstruction / cryo EM / Resolution: 20.88 Å | |||||||||
 Authors Authors | Videbaek TE / Rogers WB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Economical routes to size-specific assembly of self-closing structures. Authors: Thomas E Videbæk / Daichi Hayakawa / Gregory M Grason / Michael F Hagan / Seth Fraden / W Benjamin Rogers /  Abstract: Programmable self-assembly has seen an explosion in the diversity of synthetic crystalline materials, but developing strategies that target "self-limiting" assemblies has remained a challenge. Among ...Programmable self-assembly has seen an explosion in the diversity of synthetic crystalline materials, but developing strategies that target "self-limiting" assemblies has remained a challenge. Among these, self-closing structures, in which the local curvature defines the finite global size, are prone to polymorphism due to thermal bending fluctuations, a problem that worsens with increasing target size. Here, we show that assembly complexity can be used to eliminate this source of polymorphism in the assembly of tubules. Using many distinct components, we prune the local density of off-target geometries, increasing the selectivity of the tubule width and helicity to nearly 100%. We further show that by reducing the design constraints to target either the pitch or the width alone, fewer components are needed to reach complete selectivity. Combining experiments with theory, we reveal an economical limit, which determines the minimum number of components required to create arbitrary assembly sizes with full selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43227.map.gz emd_43227.map.gz | 162.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43227-v30.xml emd-43227-v30.xml emd-43227.xml emd-43227.xml | 14.2 KB 14.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43227_fsc.xml emd_43227_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_43227.png emd_43227.png | 62.7 KB | ||
| Masks |  emd_43227_msk_1.map emd_43227_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43227.cif.gz emd-43227.cif.gz | 4 KB | ||
| Others |  emd_43227_half_map_1.map.gz emd_43227_half_map_1.map.gz emd_43227_half_map_2.map.gz emd_43227_half_map_2.map.gz | 140.7 MB 140.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43227 http://ftp.pdbj.org/pub/emdb/structures/EMD-43227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43227 | HTTPS FTP |
-Validation report
| Summary document |  emd_43227_validation.pdf.gz emd_43227_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43227_full_validation.pdf.gz emd_43227_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_43227_validation.xml.gz emd_43227_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_43227_validation.cif.gz emd_43227_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43227 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43227 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43227 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43227.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43227.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map of the (10,0) monomer. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.023 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43227_msk_1.map emd_43227_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of the structure.
| File | emd_43227_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the structure. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map of the structure.
| File | emd_43227_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map of the structure. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DNA origami colloid for self-assembly of tubules: (10,0) monomer
| Entire | Name: DNA origami colloid for self-assembly of tubules: (10,0) monomer |
|---|---|
| Components |
|
-Supramolecule #1: DNA origami colloid for self-assembly of tubules: (10,0) monomer
| Supramolecule | Name: DNA origami colloid for self-assembly of tubules: (10,0) monomer type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 5.2 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK I |
- Electron microscopy
Electron microscopy
| Microscope | TFS TUNDRA |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number real images: 775 / Average exposure time: 1.4 sec. / Average electron dose: 69.8 e/Å2 |
| Electron beam | Acceleration voltage: 100 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)