[English] 日本語
 Yorodumi
Yorodumi- EMDB-43043: Myxococcus xanthus HEnc-K417N(A) protein shell with D5 symmetry (... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Myxococcus xanthus HEnc-K417N(A) protein shell with D5 symmetry (12 pentamers, 15 hexamers) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | encapsulin / nanocompartment / nanoparticle / cage / STRUCTURAL PROTEIN | |||||||||
| Biological species |  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) | |||||||||
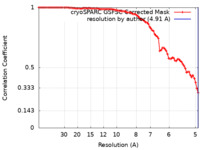
| Method | single particle reconstruction / cryo EM / Resolution: 4.91 Å | |||||||||
 Authors Authors | Hernandez C / Jenkins MC / Kopylov M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Heterologous Prime-Boost with Immunologically Orthogonal Protein Nanoparticles for Peptide Immunofocusing. Authors: Sonia Bhattacharya / Matthew C Jenkins / Parisa Keshavarz-Joud / Alisyn Retos Bourque / Keiyana White / Amina M Alvarez Barkane / Anton V Bryksin / Carolina Hernandez / Mykhailo Kopylov / M G Finn /  Abstract: Protein nanoparticles are effective platforms for antigen presentation and targeting effector immune cells in vaccine development. Encapsulins are a class of protein-based microbial nanocompartments ...Protein nanoparticles are effective platforms for antigen presentation and targeting effector immune cells in vaccine development. Encapsulins are a class of protein-based microbial nanocompartments that self-assemble into icosahedral structures with external diameters ranging from 24 to 42 nm. Encapsulins from were designed to package bacterial RNA when produced in and were shown to have immunogenic and self-adjuvanting properties enhanced by this RNA. We genetically incorporated a 20-mer peptide derived from a mutant strain of the SARS-CoV-2 receptor binding domain (RBD) into the encapsulin protomeric coat protein for presentation on the exterior surface of the particle. This immunogen elicited conformationally-relevant humoral responses to the SARS-CoV-2 RBD. Immunological recognition was enhanced when the same peptide was presented in a heterologous prime/boost vaccination strategy using the engineered encapsulin and a previously reported variant of the PP7 virus-like particle, leading to the development of a selective antibody response against a SARS-CoV-2 RBD point mutant. While generating epitope-focused antibody responses is an interplay between inherent vaccine properties and B/T cells, here we demonstrate the use of orthogonal nanoparticles to fine-tune the control of epitope focusing. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43043.map.gz emd_43043.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43043-v30.xml emd-43043-v30.xml emd-43043.xml emd-43043.xml | 12.9 KB 12.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43043_fsc.xml emd_43043_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_43043.png emd_43043.png | 104.4 KB | ||
| Masks |  emd_43043_msk_1.map emd_43043_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-43043.cif.gz emd-43043.cif.gz | 4.2 KB | ||
| Others |  emd_43043_half_map_1.map.gz emd_43043_half_map_1.map.gz emd_43043_half_map_2.map.gz emd_43043_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43043 http://ftp.pdbj.org/pub/emdb/structures/EMD-43043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43043 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43043 | HTTPS FTP |
-Validation report
| Summary document |  emd_43043_validation.pdf.gz emd_43043_validation.pdf.gz | 926.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43043_full_validation.pdf.gz emd_43043_full_validation.pdf.gz | 926 KB | Display | |
| Data in XML |  emd_43043_validation.xml.gz emd_43043_validation.xml.gz | 16.5 KB | Display | |
| Data in CIF |  emd_43043_validation.cif.gz emd_43043_validation.cif.gz | 21.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43043 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43043 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43043 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43043 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43043.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43043.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.408 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_43043_msk_1.map emd_43043_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_43043_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_43043_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Myxococcus xanthus EncA protein modified with a his-tag and K417N...
| Entire | Name: Myxococcus xanthus EncA protein modified with a his-tag and K417N(A) insertion: icosahedral T1 shell |
|---|---|
| Components |
|
-Supramolecule #1: Myxococcus xanthus EncA protein modified with a his-tag and K417N...
| Supramolecule | Name: Myxococcus xanthus EncA protein modified with a his-tag and K417N(A) insertion: icosahedral T1 shell type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Myxococcus xanthus (bacteria) Myxococcus xanthus (bacteria) |
| Molecular weight | Theoretical: 5.078 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4.2 mg/mL |
|---|---|
| Buffer | pH: 8.5 |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV / Details: 10 second wait, 1 second blot. |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 2971 / Average exposure time: 1.0 sec. / Average electron dose: 50.0 e/Å2 Details: NYSBC session m23aug21f, px size 1.204 A, 25 frames |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)