[English] 日本語
 Yorodumi
Yorodumi- EMDB-42977: Cryo-EM structure of singly-bound SNF2h-nucleosome complex with S... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | |||||||||||||||
 Map data Map data | Single-bound SNF2h nucleosome complex variability component 2A (cryoSPARC non-uniform refinement map) | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nucleosome / chromatin remodeler / ISWI / SNF2h / DNA BINDING PROTEIN-DNA complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationRSF complex / histone octamer slider activity / ACF complex / WICH complex / negative regulation of mitotic chromosome condensation / CERF complex / CHRAC / NoRC complex / NURF complex / nucleosome array spacer activity ...RSF complex / histone octamer slider activity / ACF complex / WICH complex / negative regulation of mitotic chromosome condensation / CERF complex / CHRAC / NoRC complex / NURF complex / nucleosome array spacer activity / B-WICH complex / rDNA heterochromatin formation / ATP-dependent chromatin remodeler activity / chromatin silencing complex / negative regulation of transcription by RNA polymerase I / positive regulation of transcription by RNA polymerase III / DNA methylation-dependent constitutive heterochromatin formation / positive regulation of transcription by RNA polymerase I / regulation of DNA replication / pericentric heterochromatin / nucleosome binding / condensed chromosome / Deposition of new CENPA-containing nucleosomes at the centromere / antiviral innate immune response / positive regulation of DNA replication / cellular response to leukemia inhibitory factor / DNA-templated transcription initiation / helicase activity / NoRC negatively regulates rRNA expression / B-WICH complex positively regulates rRNA expression / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / fibrillar center / structural constituent of chromatin / heterochromatin formation / nucleosome / nucleosome assembly / Recruitment and ATM-mediated phosphorylation of repair and signaling proteins at DNA double strand breaks / site of double-strand break / chromatin organization / chromatin remodeling / protein heterodimerization activity / DNA repair / DNA damage response / chromatin binding / regulation of DNA-templated transcription / regulation of transcription by RNA polymerase II / chromatin / nucleolus / positive regulation of transcription by RNA polymerase II / ATP hydrolysis activity / DNA binding / nucleoplasm / ATP binding / nucleus Similarity search - Function | |||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) / | |||||||||||||||
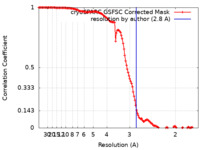
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||
 Authors Authors | Chio US / Palovcak E / Armache JP / Narlikar GJ / Cheng Y | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking. Authors: Un Seng Chio / Eugene Palovcak / Anton A A Smith / Henriette Autzen / Elise N Muñoz / Zanlin Yu / Feng Wang / David A Agard / Jean-Paul Armache / Geeta J Narlikar / Yifan Cheng /   Abstract: Single-particle cryo-EM is widely used to determine enzyme-nucleosome complex structures. However, cryo-EM sample preparation remains challenging and inconsistent due to complex denaturation at the ...Single-particle cryo-EM is widely used to determine enzyme-nucleosome complex structures. However, cryo-EM sample preparation remains challenging and inconsistent due to complex denaturation at the air-water interface (AWI). Here, to address this issue, we develop graphene-oxide-coated EM grids functionalized with either single-stranded DNA (ssDNA) or thiol-poly(acrylic acid-co-styrene) (TAASTY) co-polymer. These grids protect complexes between the chromatin remodeler SNF2h and nucleosomes from the AWI and facilitate collection of high-quality micrographs of intact SNF2h-nucleosome complexes in the absence of crosslinking. The data yields maps ranging from 2.3 to 3 Å in resolution. 3D variability analysis reveals nucleotide-state linked conformational changes in SNF2h bound to a nucleosome. In addition, the analysis provides structural evidence for asymmetric coordination between two SNF2h protomers acting on the same nucleosome. We envision these grids will enable similar detailed structural analyses for other enzyme-nucleosome complexes and possibly other protein-nucleic acid complexes in general. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42977.map.gz emd_42977.map.gz | 55.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42977-v30.xml emd-42977-v30.xml emd-42977.xml emd-42977.xml | 31.3 KB 31.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42977_fsc.xml emd_42977_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42977.png emd_42977.png | 125.2 KB | ||
| Filedesc metadata |  emd-42977.cif.gz emd-42977.cif.gz | 8.5 KB | ||
| Others |  emd_42977_half_map_1.map.gz emd_42977_half_map_1.map.gz emd_42977_half_map_2.map.gz emd_42977_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42977 http://ftp.pdbj.org/pub/emdb/structures/EMD-42977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42977 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42977 | HTTPS FTP |
-Validation report
| Summary document |  emd_42977_validation.pdf.gz emd_42977_validation.pdf.gz | 937.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42977_full_validation.pdf.gz emd_42977_full_validation.pdf.gz | 937.6 KB | Display | |
| Data in XML |  emd_42977_validation.xml.gz emd_42977_validation.xml.gz | 21.2 KB | Display | |
| Data in CIF |  emd_42977_validation.cif.gz emd_42977_validation.cif.gz | 27.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42977 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42977 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42977 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42977 | HTTPS FTP |
-Related structure data
| Related structure data |  8v4yMC  8v6vC  8v7lC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42977.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42977.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-bound SNF2h nucleosome complex variability component 2A (cryoSPARC non-uniform refinement map) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8468 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map B
| File | emd_42977_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_42977_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : ATP-dependent chromatin remodeler SNF2h in complex with nucleosome
+Supramolecule #1: ATP-dependent chromatin remodeler SNF2h in complex with nucleosome
+Supramolecule #2: Nucleosome with Xenopus laevis histones
+Supramolecule #3: SNF2h
+Macromolecule #1: Histone H3.2
+Macromolecule #2: Histone H4
+Macromolecule #3: Histone H2A type 1
+Macromolecule #4: Histone H2B
+Macromolecule #7: SWI/SNF-related matrix-associated actin-dependent regulator of ch...
+Macromolecule #5: Widom 601 DNA (147-mer) with 60 base pairs flanking DNA (reverse ...
+Macromolecule #6: Widom 601 DNA (147-mer) with 60 base pairs flanking DNA (forward ...
+Macromolecule #8: ADENOSINE-5'-DIPHOSPHATE
+Macromolecule #9: MAGNESIUM ION
+Macromolecule #10: BERYLLIUM TRIFLUORIDE ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Component:
Details: 12.5 mM HEPES-KOH, pH 7.5, 60 mM KCl, 5 mM MgCl2, 2 mM ADP, 2 mM BeSO4, 10 mM NaF, 1.5% glycerol | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 1 | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||||||||||||||
| Details | 100 nM nucleosome with 500 nM SNF2h |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)