+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | PNMA2 capsid, overall icosahedral map | |||||||||
 Map data Map data | PNMA2 capsid, overall refinement, I4 symmetry | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | capsid / endogenous retrovirus-like particle / paraneoplasm / antigen / VIRUS LIKE PARTICLE | |||||||||
| Function / homology | Paraneoplastic antigen Ma / : / : / PNMA / PNMA N-terminal RRM-like domain / positive regulation of apoptotic process / nucleolus / Paraneoplastic antigen Ma2 Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Wilkinson ME / Madigan V / Zhang Y / Zhang F | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Human paraneoplastic antigen Ma2 (PNMA2) forms icosahedral capsids that can be engineered for mRNA delivery. Authors: Victoria Madigan / Yugang Zhang / Rumya Raghavan / Max E Wilkinson / Guilhem Faure / Elena Puccio / Michael Segel / Blake Lash / Rhiannon K Macrae / Feng Zhang /  Abstract: A number of endogenous genes in the human genome encode retroviral -like proteins, which were domesticated from ancient retroelements. The paraneoplastic Ma antigen (PNMA) family members encode a - ...A number of endogenous genes in the human genome encode retroviral -like proteins, which were domesticated from ancient retroelements. The paraneoplastic Ma antigen (PNMA) family members encode a -like capsid domain, but their ability to assemble as capsids and traffic between cells remains mostly uncharacterized. Here, we systematically investigate human PNMA proteins and find that a number of PNMAs are secreted by human cells. We determine that PNMA2 forms icosahedral capsids efficiently but does not naturally encapsidate nucleic acids. We resolve the cryoelectron microscopy (cryo-EM) structure of PNMA2 and leverage the structure to design engineered PNMA2 (ePNMA2) particles with RNA packaging abilities. Recombinantly purified ePNMA2 proteins package mRNA molecules into icosahedral capsids and can function as delivery vehicles in mammalian cell lines, demonstrating the potential for engineered endogenous capsids as a nucleic acid therapy delivery modality. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42812.map.gz emd_42812.map.gz | 165.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42812-v30.xml emd-42812-v30.xml emd-42812.xml emd-42812.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42812_fsc.xml emd_42812_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_42812.png emd_42812.png | 189.5 KB | ||
| Masks |  emd_42812_msk_1.map emd_42812_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42812.cif.gz emd-42812.cif.gz | 6 KB | ||
| Others |  emd_42812_half_map_1.map.gz emd_42812_half_map_1.map.gz emd_42812_half_map_2.map.gz emd_42812_half_map_2.map.gz | 140 MB 140 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42812 http://ftp.pdbj.org/pub/emdb/structures/EMD-42812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42812 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42812 | HTTPS FTP |
-Validation report
| Summary document |  emd_42812_validation.pdf.gz emd_42812_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42812_full_validation.pdf.gz emd_42812_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42812_validation.xml.gz emd_42812_validation.xml.gz | 20.1 KB | Display | |
| Data in CIF |  emd_42812_validation.cif.gz emd_42812_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42812 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42812 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42812 | HTTPS FTP |
-Related structure data
| Related structure data |  8uyoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42812.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42812.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PNMA2 capsid, overall refinement, I4 symmetry | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.9654 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42812_msk_1.map emd_42812_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
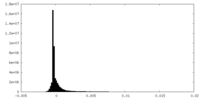
| Density Histograms |
-Half map: Refinement half-map 1
| File | emd_42812_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half-map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
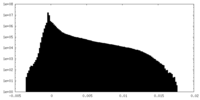
| Density Histograms |
-Half map: Refinement half-map 2
| File | emd_42812_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement half-map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
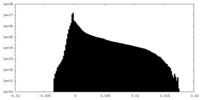
| Density Histograms |
- Sample components
Sample components
-Entire : PNMA2 icosahedral capsid
| Entire | Name: PNMA2 icosahedral capsid |
|---|---|
| Components |
|
-Supramolecule #1: PNMA2 icosahedral capsid
| Supramolecule | Name: PNMA2 icosahedral capsid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 2.49 MDa |
-Macromolecule #1: Paraneoplastic antigen Ma2
| Macromolecule | Name: Paraneoplastic antigen Ma2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.557289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MALALLEDWC RIMSVDEQKS LMVTGIPADF EEAEIQEVLQ ETLKSLGRYR LLGKIFRKQE NANAVLLELL EDTDVSAIPS EVQGKGGVW KVIFKTPNQD TEFLERLNLF LEKEGQTVSG MFRALGQEGV SPATVPCISP ELLAHLLGQA MAHAPQPLLP M RYRKLRVF ...String: MALALLEDWC RIMSVDEQKS LMVTGIPADF EEAEIQEVLQ ETLKSLGRYR LLGKIFRKQE NANAVLLELL EDTDVSAIPS EVQGKGGVW KVIFKTPNQD TEFLERLNLF LEKEGQTVSG MFRALGQEGV SPATVPCISP ELLAHLLGQA MAHAPQPLLP M RYRKLRVF SGSAVPAPEE ESFEVWLEQA TEIVKEWPVT EAEKKRWLAE SLRGPALDLM HIVQADNPSI SVEECLEAFK QV FGSLESR RTAQVRYLKT YQEEGEKVSA YVLRLETLLR RAVEKRAIPR RIADQVRLEQ VMAGATLNQM LWCRLRELKD QGP PPSFLE LMKVIREEEE EEASFENESI EEPEERDGYG RWNHEGDD UniProtKB: Paraneoplastic antigen Ma2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: phosphate buffered saline, pH 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 17600 / Average exposure time: 0.6 sec. / Average electron dose: 30.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)