+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Acinetobacter baumannii Tse15 Rhs effector | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rhs-effector / Rhs cargo effector / Type 6 secretion system effector / Acinetobacter baumannii toxin / Type VI secretion system / YD-repeat protein / T6SS. / TOXIN | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) | |||||||||
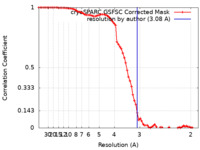
| Method | single particle reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Hayes BK / Venugopal H / McGowan S | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery. Authors: Brooke K Hayes / Marina Harper / Hariprasad Venugopal / Jessica M Lewis / Amy Wright / Han-Chung Lee / Joel R Steele / David L Steer / Ralf B Schittenhelm / John D Boyce / Sheena McGowan /  Abstract: The type VI secretion system (T6SS) is a molecular machine utilised by many Gram-negative bacteria to deliver antibacterial toxins into adjacent cells. Here we present the structure of Tse15, a T6SS ...The type VI secretion system (T6SS) is a molecular machine utilised by many Gram-negative bacteria to deliver antibacterial toxins into adjacent cells. Here we present the structure of Tse15, a T6SS Rhs effector from the nosocomial pathogen Acinetobacter baumannii. Tse15 forms a triple layered β-cocoon Rhs domain with an N-terminal α-helical clade domain and an unfolded C-terminal toxin domain inside the Rhs cage. Tse15 is cleaved into three domains, through independent auto-cleavage events involving aspartyl protease activity for toxin self-cleavage and a nucleophilic glutamic acid for N-terminal clade cleavage. Proteomic analyses identified that significantly more peptides from the N-terminal clade and toxin domains were secreted than from the Rhs cage, suggesting toxin delivery often occurs without the cage. We propose the clade domain acts as an internal chaperone to mediate toxin tethering to the T6SS machinery. Conservation of the clade domain in other Gram-negative bacteria suggests this may be a common mechanism for delivery. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42792.map.gz emd_42792.map.gz | 21.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42792-v30.xml emd-42792-v30.xml emd-42792.xml emd-42792.xml | 24.5 KB 24.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42792_fsc.xml emd_42792_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_42792.png emd_42792.png | 47.9 KB | ||
| Masks |  emd_42792_msk_1.map emd_42792_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42792.cif.gz emd-42792.cif.gz | 7.4 KB | ||
| Others |  emd_42792_additional_1.map.gz emd_42792_additional_1.map.gz emd_42792_half_map_1.map.gz emd_42792_half_map_1.map.gz emd_42792_half_map_2.map.gz emd_42792_half_map_2.map.gz | 19.6 MB 39.8 MB 39.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42792 http://ftp.pdbj.org/pub/emdb/structures/EMD-42792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42792 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42792 | HTTPS FTP |
-Validation report
| Summary document |  emd_42792_validation.pdf.gz emd_42792_validation.pdf.gz | 805 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42792_full_validation.pdf.gz emd_42792_full_validation.pdf.gz | 804.5 KB | Display | |
| Data in XML |  emd_42792_validation.xml.gz emd_42792_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_42792_validation.cif.gz emd_42792_validation.cif.gz | 19.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42792 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42792 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42792 | HTTPS FTP |
-Related structure data
| Related structure data |  8uy4MC  8uxtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42792.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42792.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.94 Å | ||||||||||||||||||||||||||||||||||||
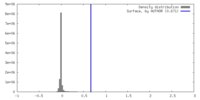
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42792_msk_1.map emd_42792_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
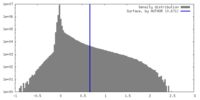
| Density Histograms |
-Additional map: Difference map generated in Chimera after refinement of...
| File | emd_42792_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Difference map generated in Chimera after refinement of A-chain. Used to guide modelling of unsequenced toxin in chains B, C, D, E. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_42792_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_42792_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Recombinant Tse15
| Entire | Name: Recombinant Tse15 |
|---|---|
| Components |
|
-Supramolecule #1: Recombinant Tse15
| Supramolecule | Name: Recombinant Tse15 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 183 KDa |
-Macromolecule #1: Tse15
| Macromolecule | Name: Tse15 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 182.818422 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAWSHPQFEK SAAKENTQQK EIKRQPAVEL AVFNLNSVTD VADLQMIASQ VQLYLQVCGN TTLEQIKSKA NITTVANIFA LTGSVLDLM LYATDKKTGD AAVQRGALLA ANLIGLFSEP NNEAHARMAL RPMFGLMAEC LYRENGKIKE TDIKRLGLHL N AMIAGDLE ...String: MAWSHPQFEK SAAKENTQQK EIKRQPAVEL AVFNLNSVTD VADLQMIASQ VQLYLQVCGN TTLEQIKSKA NITTVANIFA LTGSVLDLM LYATDKKTGD AAVQRGALLA ANLIGLFSEP NNEAHARMAL RPMFGLMAEC LYRENGKIKE TDIKRLGLHL N AMIAGDLE NFLKETQAKL SSLLISATTL GVTILQSMAT PATGINAGIT TAAGASAEKR DPKLKFTNWA VPLIDLLGKP SQ ANLTPKI QPNITSRLQQ EATQAIAALS QTLQQQANAG QKYTLAWLLQ ETLKAIQALE NKGNASVPIN QTGEYERHTK GDT LEFVSL QADALNAPPC EGADSQSGKS ISYSIGAERV QHADFYLPKI GFSFIRQYNS QMDEFDQSMV GARWMMPFSN MIQQ NAQGY LFIDSKGRKH QLPVSIIFET YEVPYEGWII KPLKNGELIL DFGGEWRSHF QSFDGGKNYY LVKKMNETSQ EEILL EYLL LDHIAYLKVI NFKLKQAEYE LKFAFNEQVK IIAVFLDDKA EPLARYEYDT QGNLIKAIDQ NGHTRTYEYN QFHQLT RYT DRTGRGQNIR YESTEAKAKA IEEWADDGSF HTKLKWHPRL RQVAVYDAYD VPTYYYFDLD GFTYRTRLAD GRESWYS RD GKKRITRQID FDGRETQQEY NDQDQLVKIV QPNGGIIRFA YNKQGNLVEI KDPEGSIWKR EYDENRNVSK EINPLGHI T QYKYNNDNQL VEVIDAKGGV KKIQYNELGQ MISYTDCSGK SSTWEYDEDG ALTAEQTANN KVVQYFYSTK GRDKGQLQS IIYPDGLKEY FEHDEEGRLL KHTDTKGLVT EYKYNQVGLL EQRIDANRHS VAYQWDKQGR IQKLINQNQA EYLFGYNPYG YLIREQAFD GEEKHYSYNE NGRLFQIRRP NILTQFDYYA DGQIASKSFT HLHTGQKQTE QFDYNLNSQL SRASNEVSQI D LYRNALGQ LVREHQHYKI PELKPLTAVL HYEYDELGNL IKTIRPDGHT LNHLVYGSGH IYAIGLNNQE VVSFQRDDLH RE TTRLLAN GLMQTKQYND VGLLSSQFIQ PEQETQDYLQ YQAHRKYHYD KNYLLSQVED SRLGKLNYQY DPIGRLIAAQ SLH KTESFN FDPAGNLIDS ESVLSPAQIK NNLIKSYKGK HYQYDVQGNV TEIIQAGKNL KLTWDNQNRL IRSDNNGLVT EYGY DVFGR RLYKKTAKEL TLFGWDGDLM IWESFKSAQT NYTKHYIYEP DSFVPLLQAG YKDFIQLIET PDYQEYQTKP YSIYK DPVW NRNLGKERTA LEQFTFYHCD QVGTPQTMTN IRGECVWEIL QDTWGAVSQI KALNQDNPFE QNNLRFQGQY YDRETE LHY NRYRYYEPHS ARYVSKDPIG LEGGMNTSSY VSDPNQWIDP KGLNSFNYGE MFGIPASAQS GLAYQGQRNY ECYAETG EL CKIKVPPLFD YVACSGGGLG IGVGFVKNQW TGEYYISGSK DSLLIPVAKS VAQNKQFSAK DLAGASCVGG NIHNIPSY T KTTMTMGEIT NEFVSGASVT VGGGAYGAVA NVVVPLVSKS SPVKGTWASE LGVGTPGFNV GVSGTVSVDT ILDAVKPSK KHHHHHH UniProtKB: Tse15 |
-Macromolecule #2: Tse15 toxin peptide (polyUNK)
| Macromolecule | Name: Tse15 toxin peptide (polyUNK) / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 2.486056 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #3: Tse15 toxin peptide (polyUNK)
| Macromolecule | Name: Tse15 toxin peptide (polyUNK) / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 954.168 Da |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) |
-Macromolecule #4: Tse15 toxin peptide (polyUNK)
| Macromolecule | Name: Tse15 toxin peptide (polyUNK) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 3.762629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) |
-Macromolecule #5: Tse15 toxin peptide (polyUNK)
| Macromolecule | Name: Tse15 toxin peptide (polyUNK) / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acinetobacter baumannii AB307-0294 (bacteria) Acinetobacter baumannii AB307-0294 (bacteria) |
| Molecular weight | Theoretical: 2.060531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 50 mM Hepes pH 8.0; 300 mM NaCl |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 150000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Initial fitting with done using UCSF Chimera using fit map. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-8uy4: |
 Movie
Movie Controller
Controller



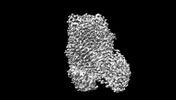

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)