+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Pr state of Stigmatella aurantiaca bacteriophytochrome 2 | |||||||||
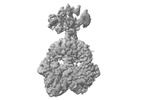
 Map data Map data | Pr state of Stigmatella aurantiaca bacteriophytochrome 2 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Red light photoreceptor / two component system / SIGNALING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdetection of visible light / histidine kinase / phosphorelay sensor kinase activity / photoreceptor activity / regulation of DNA-templated transcription Similarity search - Function | |||||||||
| Biological species |  Stigmatella aurantiaca (bacteria) Stigmatella aurantiaca (bacteria) | |||||||||
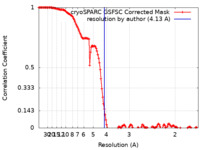
| Method | single particle reconstruction / cryo EM / Resolution: 4.13 Å | |||||||||
 Authors Authors | Malla TN / Schmidt M / Stojkovic EA | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: Photoreception and signaling in bacterial phytochrome revealed by single-particle cryo-EM. Authors: Tek Narsingh Malla / Carolina Hernandez / Srinivasan Muniyappan / David Menendez / Dorina Bizhga / Joshua H Mendez / Peter Schwander / Emina A Stojković / Marius Schmidt /  Abstract: Phytochromes are red-light photoreceptors discovered in plants with homologs in bacteria and fungi that regulate a variety of physiological responses. They display a reversible photocycle between two ...Phytochromes are red-light photoreceptors discovered in plants with homologs in bacteria and fungi that regulate a variety of physiological responses. They display a reversible photocycle between two distinct states: a red-light-absorbing Pr state and a far-red light-absorbing Pfr state. The photoconversion regulates the activity of an enzymatic domain, usually a histidine kinase (HK). The molecular mechanism that explains how light controls the HK activity is not understood because structures of unmodified bacterial phytochromes with HK activity are missing. Here, we report three cryo-electron microscopy structures of a wild-type bacterial phytochrome with HK activity determined as Pr and Pfr homodimers and as a Pr/Pfr heterodimer with individual subunits in distinct states. We propose that the Pr/Pfr heterodimer is a physiologically relevant signal transduction intermediate. Our results offer insight into the molecular mechanism that controls the enzymatic activity of the HK as part of a bacterial two-component system that perceives and transduces light signals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42448.map.gz emd_42448.map.gz | 120.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42448-v30.xml emd-42448-v30.xml emd-42448.xml emd-42448.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42448_fsc.xml emd_42448_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_42448.png emd_42448.png | 39.6 KB | ||
| Filedesc metadata |  emd-42448.cif.gz emd-42448.cif.gz | 5.8 KB | ||
| Others |  emd_42448_half_map_1.map.gz emd_42448_half_map_1.map.gz emd_42448_half_map_2.map.gz emd_42448_half_map_2.map.gz | 226.3 MB 226.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42448 http://ftp.pdbj.org/pub/emdb/structures/EMD-42448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42448 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42448 | HTTPS FTP |
-Validation report
| Summary document |  emd_42448_validation.pdf.gz emd_42448_validation.pdf.gz | 837.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42448_full_validation.pdf.gz emd_42448_full_validation.pdf.gz | 837.4 KB | Display | |
| Data in XML |  emd_42448_validation.xml.gz emd_42448_validation.xml.gz | 21.9 KB | Display | |
| Data in CIF |  emd_42448_validation.cif.gz emd_42448_validation.cif.gz | 28.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42448 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42448 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42448 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42448 | HTTPS FTP |
-Related structure data
| Related structure data |  8uphMC  8upkC  8upmC  8uqiC  8uqkC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42448.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42448.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pr state of Stigmatella aurantiaca bacteriophytochrome 2 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.844 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Stigmatella aurantiaca bacteriophytochrome
| Entire | Name: Stigmatella aurantiaca bacteriophytochrome |
|---|---|
| Components |
|
-Supramolecule #1: Stigmatella aurantiaca bacteriophytochrome
| Supramolecule | Name: Stigmatella aurantiaca bacteriophytochrome / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Stigmatella aurantiaca (bacteria) Stigmatella aurantiaca (bacteria) |
-Macromolecule #1: Bacteriophytochrome (Light-regulated signal transduction histidin...
| Macromolecule | Name: Bacteriophytochrome (Light-regulated signal transduction histidine kinase) type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Stigmatella aurantiaca (bacteria) Stigmatella aurantiaca (bacteria) |
| Molecular weight | Theoretical: 81.150758 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRPSVSELEL SQCDREPIHL LGGIQAYGVL LAFRGPDRRL EVVSANTQAL LGRPPEALLG KTAAQVLPAE LWAQWELLSA RGALRVALP SGPYRALLHE SDGLTVLELE PAELQPDMEE TALELVRRLV SPLAGAKGTR ELLQTAANTV RALTGFDRVM V YRFDADWH ...String: MRPSVSELEL SQCDREPIHL LGGIQAYGVL LAFRGPDRRL EVVSANTQAL LGRPPEALLG KTAAQVLPAE LWAQWELLSA RGALRVALP SGPYRALLHE SDGLTVLELE PAELQPDMEE TALELVRRLV SPLAGAKGTR ELLQTAANTV RALTGFDRVM V YRFDADWH GEVLAESKRE GMDGFLGMHF PATDIPVQAR ALYTRNPLRL IANARARPVP LVPSVVPELG RPLDLSGSAL RS VSPIHLE YLRNMGVEAS FSLSLLKDGA LWGLIACHHL APLHLSYERR RACEVLTQLL ALQLSSEERA AEAAEDSHRA ALL GPLATS LGAGGTLEEA LAKDGARVLE LTGATGAALL LGGEPLLVGR TPSMDEVEAL AAWLAPQPFQ TSFHTERLGS LYPP LAARA DVAAGLLAVR LAPASSRLAL WFRPEAVRTI SWAGNPRKPA EPEPGHARLH PRGSFQAWEE TVRETSPAWK RADLA AAEG FRSALIGVVL RQAAELERLS EALSRSNAEL DAFGHTVAHD LKEPLRGIQQ YAGFVMEDYH GALGPEGRGH MESLMW LAQ RSGDMLDGLF EYSRAGRVDL AWGEVNMQEV VDEVLATLST RFQAEKLTVR MPRRLPTVRC DGIRIAQVWA NLLVNAA KY QEGPERWVEA GFHGPGEPRP GAAGRYASAY VFYVKDPGIG IAPPFHEAIF EMFRRLHSAK AYGGGTGVGL AIARRLVQ L HGGALWVDSA PKQGATFYFT LGRGPG UniProtKB: histidine kinase |
-Macromolecule #2: BILIVERDINE IX ALPHA
| Macromolecule | Name: BILIVERDINE IX ALPHA / type: ligand / ID: 2 / Number of copies: 2 / Formula: BLA |
|---|---|
| Molecular weight | Theoretical: 582.646 Da |
| Chemical component information |  ChemComp-BLA: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.39 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)