[English] 日本語
 Yorodumi
Yorodumi- EMDB-42378: Structure of PsCas9 in complex with gRNA and DNA in product state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of PsCas9 in complex with gRNA and DNA in product state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cas9 / CRISPR / IMMUNE SYSTEM | |||||||||
| Biological species |  Parasutterella secunda (bacteria) / synthetic construct (others) Parasutterella secunda (bacteria) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Bravo JPK / Taylor DW | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure-guided engineering of PsCas9 yields a high fidelity and activity enzyme for in vivo gene editing Authors: Bravo JPK / Taylor DW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42378.map.gz emd_42378.map.gz | 141.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42378-v30.xml emd-42378-v30.xml emd-42378.xml emd-42378.xml | 16.9 KB 16.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_42378.png emd_42378.png | 90.8 KB | ||
| Filedesc metadata |  emd-42378.cif.gz emd-42378.cif.gz | 6.6 KB | ||
| Others |  emd_42378_half_map_1.map.gz emd_42378_half_map_1.map.gz emd_42378_half_map_2.map.gz emd_42378_half_map_2.map.gz | 262 MB 262 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42378 http://ftp.pdbj.org/pub/emdb/structures/EMD-42378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42378 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42378 | HTTPS FTP |
-Validation report
| Summary document |  emd_42378_validation.pdf.gz emd_42378_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42378_full_validation.pdf.gz emd_42378_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_42378_validation.xml.gz emd_42378_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_42378_validation.cif.gz emd_42378_validation.cif.gz | 19.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42378 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42378 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42378 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42378 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42378.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42378.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||
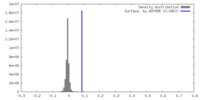
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_42378_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
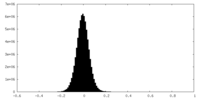
| Density Histograms |
-Half map: #2
| File | emd_42378_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
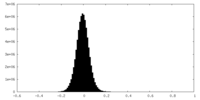
| Density Histograms |
- Sample components
Sample components
-Entire : PsCas9 in with gRNA and dsDNA, product state
| Entire | Name: PsCas9 in with gRNA and dsDNA, product state |
|---|---|
| Components |
|
-Supramolecule #1: PsCas9 in with gRNA and dsDNA, product state
| Supramolecule | Name: PsCas9 in with gRNA and dsDNA, product state / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Parasutterella secunda (bacteria) Parasutterella secunda (bacteria) |
| Molecular weight | Theoretical: 211.54 KDa |
-Macromolecule #1: Cas9
| Macromolecule | Name: Cas9 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Parasutterella secunda (bacteria) Parasutterella secunda (bacteria) |
| Molecular weight | Theoretical: 168.224266 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GAASMKRTAD GSEFESPKKK RKVTVKSTLA VDMGGRYTGI FSYTTDSGFP KAKEARAYVL NMPDNDALTY SMAARTQTRH RIRSQQRFV LARRLTYILI EGKLKRKLSP REKEAISSLL RRRGYSRLES ELDLSVLQGV ESGFFKCFLP NFDEDENLLT Q WTSLTDGY ...String: GAASMKRTAD GSEFESPKKK RKVTVKSTLA VDMGGRYTGI FSYTTDSGFP KAKEARAYVL NMPDNDALTY SMAARTQTRH RIRSQQRFV LARRLTYILI EGKLKRKLSP REKEAISSLL RRRGYSRLES ELDLSVLQGV ESGFFKCFLP NFDEDENLLT Q WTSLTDGY LQNNSDSRRQ IQIFLESSKD SKEFLTVVKS QHQDTKEYKN ALKVMRDDAE SMIEQSMFGH KHRRLYLEAI AQ DIPRDSR LKPIIEAFSG VEKFHHFIGN LSNLQLRALR WYFNDPSMKN NVFDKERLKS VLVRAYQFFH YPKDLTQQRA EVL NAYEGA TDILETLQTL NPELTIPPYE DQNNRRPPLD QTLWLSPRLL DQRYGDTWEI WVQNLLRSPL SKGIDENLDT ILIT TDRKA RLLERQSGRL IHYTSQKLYH SYVLQRLLDR TVENDAYLLK TLVSSNRGNS NEIHQAQERL TRDLGSQHIK KFLDF VRQY YDEVDKAKRG LWFIVEKPLM ERADIHPPMK NDSVILRLVG NILCVSDLVD LSFWTRKVKG QSTVRSLCTA IEKTRK EYG NSFNYLYQRA LYLQSKGKKL SAEDKDFIKL QSNVLLVSDV IAEALDIKEE QKKKFANPFS LAQLYNIIET EKSGFIS TT LAAVDENAWR NNLQGKARCV QLCADTVRPF DGALRNILDR QAYEIAKLKA EELLSTELKN QTIDLVVLLE SNQFAFSA S LAEVKKSANT AAIRQKVAKA QKRQQDRWLS KDERIKSASR GLCPYTGKNL GDKGEVDHII PRSLSMNYMG SILNSEANL IYCSQEGNQL KLNGRKKLSD LADNYLKVVF GTADRGTICK YIEKSVSELT DAKIVQFELL DRSQQDAVRH ALFLEDFSEA RRRIIRLLG KINTARVNGT QAWFAKSFIT KLRELTKEWC ANNQITLAFD LYRLDAQTVS QDYRKKFALI NKDWAKPDDK K QPIASHAI DAFCVFAAAK DKRNIANVLG VFDEVAEEQN LKTIAQLMPS EVNLISPKRK SILDKNEVGS RALMKEGIFA EH FLPILVR GDDCRIGFDW SESGSVKVKD ADKLFGVLDG LLKQSQKRSV NGFETYTVDR IKAFELLHDV FIRPCSQKML EQA EVLEKL HYITQNISVT SVYDAVNRQF KCREEILKDK DFDIKVDLGN RFGSAKGKIT LPAKREWEKL VNRSELKNLI KDKL SDKGS EKTPDGETLI YDIFRSIPVQ KLSHKATRRV WSLPKIPSIS SGVRIKRKDS NGNDIYQLYM LNDTKCKGFV VNEKG VIDW SSDLVADLYK QPTLTILNGR YLKADQYVRM DQWYEVDCGR DDVIVKMCPG TSGRRYIEIT QSKKQFEDWT GYISGS FWN YPVTIKLSSQ QIANFVKNSQ MPLLGKPRSG QITVITLGNT LKYWYCVESK NSMMNEAYQK AYLVHFNQSG GSASDYK DD DDKKRTADGS EFEPKKKRKV |
-Macromolecule #2: RNA (121-MER)
| Macromolecule | Name: RNA (121-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 41.934762 KDa |
| Sequence | String: AUGUCACCUC CAAUGACUAG GGGUUUCAGU UAUUCGUGAA AACGAAUGAA GUCACUCUUA AAGUGAGCUG AAAUCACUAA AAAUUAAGA UUGAACCCGG CUACUGACUC UGUCAUCCGG GUUUACUUAU UU |
-Macromolecule #3: DNA (80-MER)
| Macromolecule | Name: DNA (80-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 24.8169 KDa |
| Sequence | String: (DT)(DA)(DA)(DG)(DC)(DA)(DG)(DG)(DC)(DC) (DA)(DA)(DT)(DG)(DG)(DG)(DG)(DA)(DG)(DG) (DA)(DC)(DA)(DT)(DC)(DG)(DA)(DT)(DG) (DT)(DC)(DA)(DC)(DC)(DT)(DC)(DC)(DA)(DA) (DT) (DG)(DA)(DC)(DT)(DA)(DG) ...String: (DT)(DA)(DA)(DG)(DC)(DA)(DG)(DG)(DC)(DC) (DA)(DA)(DT)(DG)(DG)(DG)(DG)(DA)(DG)(DG) (DA)(DC)(DA)(DT)(DC)(DG)(DA)(DT)(DG) (DT)(DC)(DA)(DC)(DC)(DT)(DC)(DC)(DA)(DA) (DT) (DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG) (DT)(DG)(DG)(DG)(DC)(DA)(DA)(DC)(DC)(DA) (DC)(DA) (DA)(DA)(DC)(DC)(DC)(DA)(DC) (DG)(DA)(DG)(DG)(DG)(DC)(DA)(DG)(DA)(DG) (DT)(DG)(DC) |
-Macromolecule #4: DNA (80-MER)
| Macromolecule | Name: DNA (80-MER) / type: dna / ID: 4 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 24.5306 KDa |
| Sequence | String: (DG)(DC)(DA)(DC)(DT)(DC)(DT)(DG)(DC)(DC) (DC)(DT)(DC)(DG)(DT)(DG)(DG)(DG)(DT)(DT) (DT)(DG)(DT)(DG)(DG)(DT)(DT)(DG)(DC) (DC)(DC)(DA)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC) (DA)(DT)(DT)(DG)(DG)(DA) ...String: (DG)(DC)(DA)(DC)(DT)(DC)(DT)(DG)(DC)(DC) (DC)(DT)(DC)(DG)(DT)(DG)(DG)(DG)(DT)(DT) (DT)(DG)(DT)(DG)(DG)(DT)(DT)(DG)(DC) (DC)(DC)(DA)(DC)(DC)(DC)(DT)(DA)(DG)(DT) (DC) (DA)(DT)(DT)(DG)(DG)(DA)(DG)(DG) (DT)(DG)(DA)(DC)(DA)(DT)(DC)(DG)(DA)(DT) (DG)(DT) (DC)(DC)(DT)(DC)(DC)(DC)(DC) (DA)(DT)(DT)(DG)(DG)(DC)(DC)(DT)(DG)(DC) (DT)(DT)(DA) |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 7 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 80.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 77059 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)