[English] 日本語
 Yorodumi
Yorodumi- EMDB-41929: The mTORC1 cholesterol sensor LYCHOS (GPR155) - monomer with auxin -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | The mTORC1 cholesterol sensor LYCHOS (GPR155) - monomer with auxin | ||||||||||||
 Map data Map data | Final combined map, filtered to Gold-standard resolution. Not sharpened. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | PIN-FORMED / GPCR / Cholesterol / auxin / transporter / cell-growth / mTORC1 / cancer / MEMBRANE PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcellular response to cholesterol / cholesterol binding / negative regulation of BMP signaling pathway / positive regulation of TORC1 signaling / cellular response to amino acid starvation / transmembrane transport / cognition / intracellular signal transduction / lysosomal membrane / extracellular exosome Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
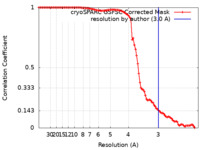
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | ||||||||||||
 Authors Authors | Bayly-Jones C / Lupton CJ / Ellisdon AM | ||||||||||||
| Funding support |  Australia, Australia,  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation | Journal: Science / Year: 2022 Title: Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Authors: Hijai R Shin / Y Rose Citron / Lei Wang / Laura Tribouillard / Claire S Goul / Robin Stipp / Yusuke Sugasawa / Aakriti Jain / Nolwenn Samson / Chun-Yan Lim / Oliver B Davis / David Castaneda- ...Authors: Hijai R Shin / Y Rose Citron / Lei Wang / Laura Tribouillard / Claire S Goul / Robin Stipp / Yusuke Sugasawa / Aakriti Jain / Nolwenn Samson / Chun-Yan Lim / Oliver B Davis / David Castaneda-Carpio / Mingxing Qian / Daniel K Nomura / Rushika M Perera / Eunyong Park / Douglas F Covey / Mathieu Laplante / Alex S Evers / Roberto Zoncu /    Abstract: Lysosomes coordinate cellular metabolism and growth upon sensing of essential nutrients, including cholesterol. Through bioinformatic analysis of lysosomal proteomes, we identified lysosomal ...Lysosomes coordinate cellular metabolism and growth upon sensing of essential nutrients, including cholesterol. Through bioinformatic analysis of lysosomal proteomes, we identified lysosomal cholesterol signaling (LYCHOS, previously annotated as G protein-coupled receptor 155), a multidomain transmembrane protein that enables cholesterol-dependent activation of the master growth regulator, the protein kinase mechanistic target of rapamycin complex 1 (mTORC1). Cholesterol bound to the amino-terminal permease-like region of LYCHOS, and mutating this site impaired mTORC1 activation. At high cholesterol concentrations, LYCHOS bound to the GATOR1 complex, a guanosine triphosphatase (GTPase)-activating protein for the Rag GTPases, through a conserved cytoplasm-facing loop. By sequestering GATOR1, LYCHOS promotes cholesterol- and Rag-dependent recruitment of mTORC1 to lysosomes. Thus, LYCHOS functions in a lysosomal pathway for cholesterol sensing and couples cholesterol concentrations to mTORC1-dependent anabolic signaling. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41929.map.gz emd_41929.map.gz | 49.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41929-v30.xml emd-41929-v30.xml emd-41929.xml emd-41929.xml | 26.7 KB 26.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41929_fsc.xml emd_41929_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41929.png emd_41929.png | 72.2 KB | ||
| Masks |  emd_41929_msk_1.map emd_41929_msk_1.map | 98.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41929.cif.gz emd-41929.cif.gz | 7.4 KB | ||
| Others |  emd_41929_additional_1.map.gz emd_41929_additional_1.map.gz emd_41929_additional_2.map.gz emd_41929_additional_2.map.gz emd_41929_half_map_1.map.gz emd_41929_half_map_1.map.gz emd_41929_half_map_2.map.gz emd_41929_half_map_2.map.gz | 93.4 MB 112 MB 91.7 MB 91.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41929 http://ftp.pdbj.org/pub/emdb/structures/EMD-41929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41929 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41929 | HTTPS FTP |
-Validation report
| Summary document |  emd_41929_validation.pdf.gz emd_41929_validation.pdf.gz | 723.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41929_full_validation.pdf.gz emd_41929_full_validation.pdf.gz | 723.4 KB | Display | |
| Data in XML |  emd_41929_validation.xml.gz emd_41929_validation.xml.gz | 18 KB | Display | |
| Data in CIF |  emd_41929_validation.cif.gz emd_41929_validation.cif.gz | 23.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41929 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41929 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41929 | HTTPS FTP |
-Related structure data
| Related structure data |  8u5nMC  8u54C  8u56C  8u58C  8u5cC  8u5qC  8u5vC  8u5xC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41929.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41929.map.gz / Format: CCP4 / Size: 98.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final combined map, filtered to Gold-standard resolution. Not sharpened. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1081 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41929_msk_1.map emd_41929_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Monomer of LYCHOS (GPR155)
| Entire | Name: Monomer of LYCHOS (GPR155) |
|---|---|
| Components |
|
-Supramolecule #1: Monomer of LYCHOS (GPR155)
| Supramolecule | Name: Monomer of LYCHOS (GPR155) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99 KDa |
-Macromolecule #1: Integral membrane protein GPR155
| Macromolecule | Name: Integral membrane protein GPR155 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.276891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MNSNLPAENL TIAVNMTKTL PTAVTHGFNS TNDPPSMSIT RLFPALLECF GIVLCGYIAG RANVITSTQA KGLGNFVSRF ALPALLFKN MVVLNFSNVD WSFLYSILIA KASVFFIVCV LTLLVASPDS RFSKAGLFPI FATQSNDFAL GYPIVEALYQ T TYPEYLQY ...String: MNSNLPAENL TIAVNMTKTL PTAVTHGFNS TNDPPSMSIT RLFPALLECF GIVLCGYIAG RANVITSTQA KGLGNFVSRF ALPALLFKN MVVLNFSNVD WSFLYSILIA KASVFFIVCV LTLLVASPDS RFSKAGLFPI FATQSNDFAL GYPIVEALYQ T TYPEYLQY IYLVAPISLM MLNPIGFIFC EIQKWKDTQN ASQNKIKIVG LGLLRVLQNP IVFMVFIGIA FNFILDRKVP VY VENFLDG LGNSFSGSAL FYLGLTMVGK IKRLKKSAFV VLILLITAKL LVLPLLCREM VELLDKGDSV VNHTSLSNYA FLY GVFPVA PGVAIFATQF NMEVEIITSG MVISTFVSAP IMYVSAWLLT FPTMDPKPLA YAIQNVSFDI SIVSLISLIW SLAI LLLSK KYKQLPHMLT TNLLIAQSIV CAGMMIWNFV KEKNFVGQIL VFVLLYSSLY STYLWTGLLA ISLFLLKKRE RVQIP VGII IISGWGIPAL LVGVLLITGK HNGDSIDSAF FYGKEQMITT AVTLFCSILI AGISLMCMNQ TAQAGSYEGF DQSQSH KVV EPGNTAFEES PAPVNEPELF TSSIPETSCC SCSMGNGELH CPSIEPIANT STSEPVIPSF EKNNHCVSRC NSQSCIL AQ EEEQYLQSGD QQLTRHVLLC LLLIIGLFAN LSSCLWWLFN QEPGRLYVEL QFFCAVFNFG QGFISFGIFG LDKHLIIL P FKRRLEFLWN NKDTAENRDS PVSEEIKMTC QQFIHYHRDL CIRNIVKERR CGAKTSAGTF CGCDLVSWLI EVGLASDRG EAVIYGDRLV QGGVIQHITN EYEFRDEYLF YRFLQKSPEQ SPPAINANTL QQERYKEIEH SSPPSHSPKT GSGSDYKDDD DKDYKDDDD K UniProtKB: Lysosomal cholesterol signaling protein |
-Macromolecule #2: 1H-INDOL-3-YLACETIC ACID
| Macromolecule | Name: 1H-INDOL-3-YLACETIC ACID / type: ligand / ID: 2 / Number of copies: 1 / Formula: IAC |
|---|---|
| Molecular weight | Theoretical: 175.184 Da |
| Chemical component information | 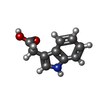 ChemComp-IAC: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number real images: 8067 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: AlphaFold / Chain - Initial model type: in silico model Details: A monomeric assembly was predicted with AlphaFold and then rigid body fit into the map. Subsequent refinements were performed in ISOLDE. |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
| Output model |  PDB-8u5n: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)