[English] 日本語
 Yorodumi
Yorodumi- EMDB-41805: Cryo-EM structure of murine Thrombopoietin receptor ectodomain in... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of murine Thrombopoietin receptor ectodomain in complex with Tpo | |||||||||
 Map data Map data | Refinement map from 3Dflex after non-uniform refinement. Sharpened with deepEMhancer | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor / cytokine / signalling / haematopoiesis / CYTOKINE-RECEPTOR complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationthrombopoietin receptor activity / regulation of chemokine production / positive regulation of hematopoietic stem cell proliferation / regulation of stem cell division / megakaryocyte differentiation / positive regulation of megakaryocyte differentiation / thrombopoietin-mediated signaling pathway / regulation of stem cell proliferation / myeloid cell differentiation / definitive hemopoiesis ...thrombopoietin receptor activity / regulation of chemokine production / positive regulation of hematopoietic stem cell proliferation / regulation of stem cell division / megakaryocyte differentiation / positive regulation of megakaryocyte differentiation / thrombopoietin-mediated signaling pathway / regulation of stem cell proliferation / myeloid cell differentiation / definitive hemopoiesis / platelet formation / homeostasis of number of cells / cytokine activity / hormone activity / positive regulation of protein phosphorylation / cell population proliferation / positive regulation of ERK1 and ERK2 cascade / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / DNA-binding transcription factor activity / positive regulation of gene expression / negative regulation of apoptotic process / cell surface / Golgi apparatus / extracellular space / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
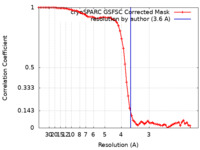
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Sarson-Lawrence KS / Hardy JM / Leis A / Babon JJ / Kershaw NJ | |||||||||
| Funding support |  Australia, 2 items Australia, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin. Authors: Kaiseal T G Sarson-Lawrence / Joshua M Hardy / Josephine Iaria / Dina Stockwell / Kira Behrens / Tamanna Saiyed / Cyrus Tan / Leila Jebeli / Nichollas E Scott / Toby A Dite / Nicos A Nicola ...Authors: Kaiseal T G Sarson-Lawrence / Joshua M Hardy / Josephine Iaria / Dina Stockwell / Kira Behrens / Tamanna Saiyed / Cyrus Tan / Leila Jebeli / Nichollas E Scott / Toby A Dite / Nicos A Nicola / Andrew P Leis / Jeffrey J Babon / Nadia J Kershaw /  Abstract: Thrombopoietin (Tpo) is the primary regulator of megakaryocyte and platelet numbers and is required for haematopoetic stem cell maintenance. Tpo functions by binding its receptor (TpoR, a homodimeric ...Thrombopoietin (Tpo) is the primary regulator of megakaryocyte and platelet numbers and is required for haematopoetic stem cell maintenance. Tpo functions by binding its receptor (TpoR, a homodimeric Class I cytokine receptor) and initiating cell proliferation or differentiation. Here we characterise the murine Tpo:TpoR signalling complex biochemically and structurally, using cryo-electron microscopy. Tpo uses opposing surfaces to recruit two copies of receptor, forming a 1:2 complex. Although it binds to the same, membrane-distal site on both receptor chains, it does so with significantly different affinities and its highly glycosylated C-terminal domain is not required. In one receptor chain, a large insertion, unique to TpoR, forms a partially structured loop that contacts cytokine. Tpo binding induces the juxtaposition of the two receptor chains adjacent to the cell membrane. The therapeutic agent romiplostim also targets the cytokine-binding site and the characterisation presented here supports the future development of improved TpoR agonists. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41805.map.gz emd_41805.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41805-v30.xml emd-41805-v30.xml emd-41805.xml emd-41805.xml | 26.4 KB 26.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41805_fsc.xml emd_41805_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41805.png emd_41805.png | 117.7 KB | ||
| Filedesc metadata |  emd-41805.cif.gz emd-41805.cif.gz | 8.2 KB | ||
| Others |  emd_41805_additional_1.map.gz emd_41805_additional_1.map.gz emd_41805_half_map_1.map.gz emd_41805_half_map_1.map.gz emd_41805_half_map_2.map.gz emd_41805_half_map_2.map.gz | 28.7 MB 28.4 MB 28.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41805 http://ftp.pdbj.org/pub/emdb/structures/EMD-41805 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41805 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41805 | HTTPS FTP |
-Validation report
| Summary document |  emd_41805_validation.pdf.gz emd_41805_validation.pdf.gz | 628.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41805_full_validation.pdf.gz emd_41805_full_validation.pdf.gz | 628.3 KB | Display | |
| Data in XML |  emd_41805_validation.xml.gz emd_41805_validation.xml.gz | 14.4 KB | Display | |
| Data in CIF |  emd_41805_validation.cif.gz emd_41805_validation.cif.gz | 18.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41805 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41805 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41805 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41805 | HTTPS FTP |
-Related structure data
| Related structure data |  8u18MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41805.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41805.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Refinement map from 3Dflex after non-uniform refinement. Sharpened with deepEMhancer | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.02626 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: 3DFlex Refinement Map
| File | emd_41805_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFlex Refinement Map | ||||||||||||
| Projections & Slices |
| ||||||||||||
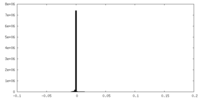
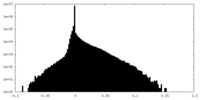
| Density Histograms |
-Half map: 3DFlex Half Map B
| File | emd_41805_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFlex Half Map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
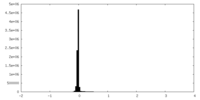
| Density Histograms |
-Half map: 3DFlex Half Map A
| File | emd_41805_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3DFlex Half Map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
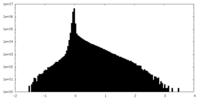
| Density Histograms |
- Sample components
Sample components
-Entire : 2:1 complex of the mouse thrombopoietin receptor ectodomain and m...
| Entire | Name: 2:1 complex of the mouse thrombopoietin receptor ectodomain and mouse thrombopoietin |
|---|---|
| Components |
|
-Supramolecule #1: 2:1 complex of the mouse thrombopoietin receptor ectodomain and m...
| Supramolecule | Name: 2:1 complex of the mouse thrombopoietin receptor ectodomain and mouse thrombopoietin type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: mTpoR was expressed as a fusion of residues 1-482 of mTPOR, followed by a GCN4 leucine zipper from Saccharomyces cerevisiae, a TEV protease site, the Fc domain of hlgG1, and a C-terminal ...Details: mTpoR was expressed as a fusion of residues 1-482 of mTPOR, followed by a GCN4 leucine zipper from Saccharomyces cerevisiae, a TEV protease site, the Fc domain of hlgG1, and a C-terminal FLAG tag. The mTpoR signal peptide (1-25) was cleaved during expression and the Fc tag was cleaved by TEV before complexation. mTPO was expressed as a fusion of the mIL3 secretion signal (cleaved during expression), an N-terminal FLAG-tag followed by residues 22 to 184 of mTPO. |
|---|---|
| Molecular weight | Theoretical: 131.698 KDa |
-Supramolecule #2: mouse thrombopoietin receptor ectodomain
| Supramolecule | Name: mouse thrombopoietin receptor ectodomain / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: mouse thrombopoietin
| Supramolecule | Name: mouse thrombopoietin / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Thrombopoietin receptor,GCN4 isoform 1
| Macromolecule | Name: Thrombopoietin receptor,GCN4 isoform 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.559676 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QDVFLLALGT EPLNCFSQTF EDLTCFWDEE EAAPSGTYQL LYAYRGEKPR ACPLYSQSVP TFGTRYVCQF PAQDEVRLFF PLHLWVKNV SLNQTLIQRV LFVDSVGLPA PPRVIKARGG SQPGELQIHW EAPAPEISDF LRHELRYGPT DSSNATAPSV I QLLSTETC ...String: QDVFLLALGT EPLNCFSQTF EDLTCFWDEE EAAPSGTYQL LYAYRGEKPR ACPLYSQSVP TFGTRYVCQF PAQDEVRLFF PLHLWVKNV SLNQTLIQRV LFVDSVGLPA PPRVIKARGG SQPGELQIHW EAPAPEISDF LRHELRYGPT DSSNATAPSV I QLLSTETC CPTLWMPNPV PVLDQPPCVH PTASQPHGPA PFLTVKGGSC LVSGLQAGKS YWLQLRSQPD GVSLRGSWGP WS FPVTVDL PGDAVTIGLQ CFTLDLKMVT CQWQQQDRTS SQGFFRHSRT RCCPTDRDPT WEKCEEEEPR PGSQPALVSR CHF KSRNDS VIHILVEVTT AQGAVHSYLG SPFWIHQAVL LPTPSLHWRE VSSGRLELEW QHQSSWAAQE TCYQLRYTGE GRED WKVLE PSLGARGGTL ELRPRARYSL QLRARLNGPT YQGPWSAWSP PARVSTGSET AWRMKQLEDK VEELLSKNYH LENEV ARLK KLVGERTGTG GAPENLYFQ UniProtKB: Thrombopoietin receptor, GCN4 isoform 1 |
-Macromolecule #2: Thrombopoietin
| Macromolecule | Name: Thrombopoietin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.611674 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ASISARQDYK DDDDKTRQSP VAPACDPRLL NKLLRDSHLL HSRLSQCPDV DPLSIPVLLP AVDFSLGEWK TQTEQSKAQD ILGAVSLLL EGVMAARGQL EPSCLSSLLG QLSGQVRLLL GALQGLLGTQ LPLQGRTTAH KDPNALFLSL QQLLRGKVRF L LLVEGPTL CVRRTLPTTA VPS UniProtKB: Thrombopoietin |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.55 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot time of 4 s, blot force of 6 and no wait time or drain time.. | |||||||||
| Details | TpoR1-479-LeuZ and TPON were combined with a molar ratio of 1:1.5 and applied to superdex 200 10/300 increased column equilibrated in TBS pH7.5. The resultant peaks were analysed by SDS-PAGE and SEC-MALS, and those corresponding to the complex were pooled and concentrated to 0.55 mg/ml. CTAB (Hexadecyl-trimethylammonium bromide) was added right before plunge freezing to a concentration of 0.005% (w/v). |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6130 / Average exposure time: 2.38 sec. / Average electron dose: 50.57 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.6 µm / Nominal defocus min: 0.4 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model Details: Multimer complex of the complete assembly was produced using AlphaFold2 on Google CoLab |
|---|---|
| Details | ChimeraX was used to perform rigid-body fitting of domains and modelling was performed using Coot and ISOLDE. Refinement was carried out in PHENIX. |
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT / Target criteria: Cross-correlation coefficient |
| Output model |  PDB-8u18: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)