+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human cytomegalovirus penton vertex, CVSC-bound configuration | ||||||||||||||||||||||||||||||
 Map data Map data | Post-processed map used for real space refinement. | ||||||||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||||||||
 Keywords Keywords | tegument / penton / CVSC / intracellular transport / VIRUS | ||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationhost cell viral assembly compartment / T=16 icosahedral viral capsid / viral tegument / viral DNA genome packaging / viral capsid assembly / host cell cytoplasmic vesicle / chromosome organization / viral process / viral penetration into host nucleus / viral capsid ...host cell viral assembly compartment / T=16 icosahedral viral capsid / viral tegument / viral DNA genome packaging / viral capsid assembly / host cell cytoplasmic vesicle / chromosome organization / viral process / viral penetration into host nucleus / viral capsid / host cell / symbiont-mediated perturbation of host ubiquitin-like protein modification / ubiquitinyl hydrolase 1 / host cell cytoplasm / cysteine-type deubiquitinase activity / Hydrolases; Acting on peptide bonds (peptidases); Cysteine endopeptidases / host cell perinuclear region of cytoplasm / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis / DNA binding Similarity search - Function | ||||||||||||||||||||||||||||||
| Biological species |   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 | ||||||||||||||||||||||||||||||
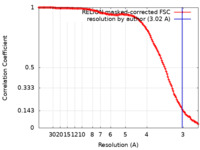
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | ||||||||||||||||||||||||||||||
 Authors Authors | Jih J / Liu YT / Liu W / Zhou H | ||||||||||||||||||||||||||||||
| Funding support |  United States, 9 items United States, 9 items
| ||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2024 Journal: Sci Adv / Year: 2024Title: The incredible bulk: Human cytomegalovirus tegument architectures uncovered by AI-empowered cryo-EM. Authors: Jonathan Jih / Yun-Tao Liu / Wei Liu / Z Hong Zhou /  Abstract: The compartmentalization of eukaryotic cells presents considerable challenges to the herpesvirus life cycle. The herpesvirus tegument, a bulky proteinaceous aggregate sandwiched between ...The compartmentalization of eukaryotic cells presents considerable challenges to the herpesvirus life cycle. The herpesvirus tegument, a bulky proteinaceous aggregate sandwiched between herpesviruses' capsid and envelope, is uniquely evolved to address these challenges, yet tegument structure and organization remain poorly characterized. We use deep-learning-enhanced cryogenic electron microscopy to investigate the tegument of human cytomegalovirus virions and noninfectious enveloped particles (NIEPs; a genome packaging-aborted state), revealing a portal-biased tegumentation scheme. We resolve atomic structures of portal vertex-associated tegument (PVAT) and identify multiple configurations of PVAT arising from layered reorganization of pUL77, pUL48 (large tegument protein), and pUL47 (inner tegument protein) assemblies. Analyses show that pUL77 seals the last-packaged viral genome end through electrostatic interactions, pUL77 and pUL48 harbor a head-linker-capsid-binding motif conducive to PVAT reconfiguration, and pUL47/48 dimers form 45-nm-long filaments extending from the portal vertex. These results provide a structural framework for understanding how herpesvirus tegument facilitates and evolves during processes spanning viral genome packaging to delivery. | ||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41204.map.gz emd_41204.map.gz | 165.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41204-v30.xml emd-41204-v30.xml emd-41204.xml emd-41204.xml | 31.1 KB 31.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41204_fsc.xml emd_41204_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_41204.png emd_41204.png | 217.4 KB | ||
| Filedesc metadata |  emd-41204.cif.gz emd-41204.cif.gz | 10.3 KB | ||
| Others |  emd_41204_additional_1.map.gz emd_41204_additional_1.map.gz emd_41204_half_map_1.map.gz emd_41204_half_map_1.map.gz emd_41204_half_map_2.map.gz emd_41204_half_map_2.map.gz | 140.3 MB 140.7 MB 140.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41204 http://ftp.pdbj.org/pub/emdb/structures/EMD-41204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41204 | HTTPS FTP |
-Validation report
| Summary document |  emd_41204_validation.pdf.gz emd_41204_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41204_full_validation.pdf.gz emd_41204_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_41204_validation.xml.gz emd_41204_validation.xml.gz | 19.9 KB | Display | |
| Data in CIF |  emd_41204_validation.cif.gz emd_41204_validation.cif.gz | 26.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41204 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41204 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41204 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41204 | HTTPS FTP |
-Related structure data
| Related structure data |  8tewMC  8tepC  8tesC  8tetC  8teuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41204.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41204.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed map used for real space refinement. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||
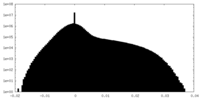
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Primary map.
| File | emd_41204_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Primary map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
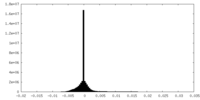
| Density Histograms |
-Half map: Half map 1.
| File | emd_41204_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
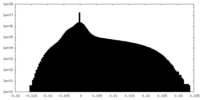
| Density Histograms |
-Half map: Half map 2.
| File | emd_41204_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
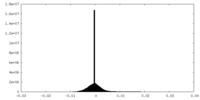
| Density Histograms |
- Sample components
Sample components
-Entire : Human herpesvirus 5 strain AD169
| Entire | Name:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
|---|---|
| Components |
|
-Supramolecule #1: Human herpesvirus 5 strain AD169
| Supramolecule | Name: Human herpesvirus 5 strain AD169 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10360 / Sci species name: Human herpesvirus 5 strain AD169 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Virion capsid / T number (triangulation number): 16 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 154.048906 KDa |
| Sequence | String: MENWSALELL PKVGIPTDFL THVKTSAGEE MFEALRIYYG DDPERYNIHF EAIFGTFCNR LEWVYFLTSG LAAAAHAIKF HDLNKLTTG KMLFHVQVPR VASGAGLPTS RQTTIMVTKY SEKSPITIPF ELSAACLTYL RETFEGTILD KILNVEAMHT V LRALKNTA ...String: MENWSALELL PKVGIPTDFL THVKTSAGEE MFEALRIYYG DDPERYNIHF EAIFGTFCNR LEWVYFLTSG LAAAAHAIKF HDLNKLTTG KMLFHVQVPR VASGAGLPTS RQTTIMVTKY SEKSPITIPF ELSAACLTYL RETFEGTILD KILNVEAMHT V LRALKNTA DAMERGLIHS FLQTLLRKAP PYFVVQTLVE NATLARQALN RIQRSNILQS FKAKMLATLF LLNRTRDRDY VL KFLTRLA EAATDSILDN PTTYTTSSGA KISGVMVSTA NVMQIIMSLL SSHITKETVS APATYGNFVL SPENAVTAIS YHS ILADFN SYKAHLTSGQ PHLPNDSLSQ AGAHSLTPLS MDVIRLGEKT VIMENLRRVY KNTDTKDPLE RNVDLTFFFP VGLY LPEDR GYTTVESKVK LNDTVRNALP TTAYLLNRDR AVQKIDFVDA LKTLCHPVLH EPAPCLQTFT ERGPPSEPAM QRLLE CRFQ QEPMGGAARR IPHFYRVRRE VPRTVNEMKQ DFVVTDFYKV GNITLYTELH PFFDFTHCQE NSETVALCTP RIVIGN LPD GLAPGPFHEL RTWEIMEHMR LRPPPDYEET LRLFKTTVTS PNYPELCYLV DVLVHGNVDA FLLIRTFVAR CIVNMFH TR QLLVFAHSYA LVTLIAEHLA DGALPPQLLF HYRNLVAVLR LVTRISALPG LNNGQLAEEP LSAYVNALHD HRLWPPFV T HLPRNMEGVQ VVADRQPLNP ANIEARHHGV SDVPRLGAMD ADEPLFVDDY RATDDEWTLQ KVFYLCLMPA MTNNRACGL GLNLKTLLVD LFYRPAFLLM PAATAVSTSG TTSKESTSGV TPEDSIAAQR QAVGEMLTEL VEDVATDAHT PLLQACRELF LAVQFVGEH VKVLEVRAPL DHAQRQGLPD FISRQHVLYN GCCVVTAPKT LIEYSLPVPF HRFYSNPTIC AALSDDIKRY V TEFPHYHR HDGGFPLPTA FAHEYHNWLR SPFSRYSATC PNVLHSVMTL AAMLYKISPV SLVLQTKAHI HPGFALTAVR TD TFEVDML LYSGKSCTSV IINNPIVTKE ERDISTTYHV TQNINTVDMG LGYTSNTCVA YVNRVRTDMG VRVQDLFRVF PMN VYRHDE VDRWIRHAAG VERPQLLDTE TISMLTFGSM SERNAAATVH GQKAACELIL TPVTMDVNYF KIPNNPRGRA SCML AVDPY DTEAATKAIY DHREADAQTF AATHNPWASQ AGCLSDVLYN TRHRERLGYN SKFYSPCAQY FNTEEIIAAN KTLFK TIDE YLLRAKDCIR GDTDTQYVCV EGTEQLIENP CRLTQEALPI LSTTTLALME TKLKGGAGAF ATSETHFGNY VVGEII PLQ QSMLFNS UniProtKB: Major capsid protein |
-Macromolecule #2: Small capsomere-interacting protein
| Macromolecule | Name: Small capsomere-interacting protein / type: protein_or_peptide / ID: 2 / Number of copies: 7 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 8.495924 KDa |
| Sequence | String: MSNTAPGPTV ANKRDEKHRH VVNVVLELPT EISEATHPVL ATMLSKYTRM SSLFNDKCAF KLDLLRMVAV SRTRR UniProtKB: Small capsomere-interacting protein |
-Macromolecule #3: Large structural phosphoprotein
| Macromolecule | Name: Large structural phosphoprotein / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 112.829102 KDa |
| Sequence | String: MSLQFIGLQR RDVVALVNFL RHLTQKPDVD LEAHPKILKK CGEKRLHRRT VLFNELMLWL GYYRELRFHN PDLSSVLEEF EVRCVAVAR RGYTYPFGDR GKARDHLAVL DRTEFDTDVR HDAEIVERAL VSAVILAKMS VRETLVTAIG QTEPIAFVHL K DTEVQRIE ...String: MSLQFIGLQR RDVVALVNFL RHLTQKPDVD LEAHPKILKK CGEKRLHRRT VLFNELMLWL GYYRELRFHN PDLSSVLEEF EVRCVAVAR RGYTYPFGDR GKARDHLAVL DRTEFDTDVR HDAEIVERAL VSAVILAKMS VRETLVTAIG QTEPIAFVHL K DTEVQRIE ENLEGVRRNM FCVKPLDLNL DRHANTALVN AVNKLVYTGR LIMNVRRSWE ELERKCLARI QERCKLLVKE LR MCLSFDS NYCRNILKHA VENGDSADTL LELLIEDFDI YVDSFPQSAH TFLGARSPSL EFDDDANLLS LGGGSAFSSV PKK HVPTQP LDGWSWIASP WKGHKPFRFE AHGSLAPAAE AHAARSAAVG YYDEEEKRRE RQKRVDDEVV QREKQQLKAW EERQ QNLQQ RQQQPPPPAR KPSASRRLFG SSADEDDDDD DDEKNIFTPI KKPGTSGKGA ASGGGVSSIF SGLLSSGSQK PTSGP LNIP QQQQRHAAFS LVSPQVTKAS PGRVRRDSAW DVRPLTETRG DLFSGDEDSD SSDGYPPNRQ DPRFTDTLVD ITDTET SAK PPVTTAYKFE QPTLTFGAGV NVPAGAGAAI LTPTPVNPST APAPAPTPTF AGTQTPVNGN SPWAPTAPLP GDMNPAN WP RERAWALKNP HLAYNPFRMP TTSTASQNTV STTPRRPSTP RAAVTQTASR DAADEVWALR DQTAESPVED SEEEDDDS S DTGSVVSLGH TTPSSDYNND VISPPSQTPE QSTPSRIRKA KLSSPMTTTS TSQKPVLGKR VATPHASARA QTVTSTPVQ GRLEKQVSGT PSTVPATLLQ PQPASSKTTS SRNVTSGAGT SSASSARQPS ASASVLSPTE DDVVSPATSP LSMLSSASPS PAKSAPPSP VKGRGSRVGV PSLKPTLGGK AVVGRPPSVP VSGSAPGRLS GSSRAASTTP TYPAVTTVYP PSSTAKSSVS N APPVASPS ILKPGASAAL QSRRSTGTAA VGSPVKSTTG MKTVAFDLSS PQKSGTGPQP GSAGMGGAKT PSDAVQNILQ KI EKIKNTE E UniProtKB: Large structural phosphoprotein |
-Macromolecule #4: Large tegument protein deneddylase
| Macromolecule | Name: Large tegument protein deneddylase / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO / EC number: ubiquitinyl hydrolase 1 |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 253.541141 KDa |
| Sequence | String: MKVTQASCHQ GDIARFGARA GNQCVCNGIM FLHALHLGGT SAVLQTEALD AIMEEGARLD ARLERELQKK LPAGGRLPVY RLGDEVPRR LESRFGRTVH ALSRPFNGTT ETCDLDGYMC PGIFDFLRYA HAKPRPTYVL VTVNSLARAV VFTEDHMLVF D PHSSAECH ...String: MKVTQASCHQ GDIARFGARA GNQCVCNGIM FLHALHLGGT SAVLQTEALD AIMEEGARLD ARLERELQKK LPAGGRLPVY RLGDEVPRR LESRFGRTVH ALSRPFNGTT ETCDLDGYMC PGIFDFLRYA HAKPRPTYVL VTVNSLARAV VFTEDHMLVF D PHSSAECH NAAVYHCEGL HQVLMVLTGF GVQLSPAFYY EALFLYMLDV ATVPEAEIAA RLVSTYRDRD IDLTGVVRES AD TAATTTT AAPSLPPLPD PIVDPGCPPG VAPSIPVYDP SSSPKKTPEK RRKDLSGSKH GGKKKPPSTT SKTLATASSS PSA IAAASS SSAVPPSYSC GEGALPALGR YQQLVDEVEQ ELKALTLPPL PANTSAWTLH AAGTESGANA ATATAPSFDE AFLT DRLQQ LIIHAVNQRS CLRRPCGPQS AAQQAVRAYL GLSKKLDAFL LNWLHHGLDL QRMHDYLSHK TTKGTYSTLD RALLE KMQV VFDPYGRQHG PALIAWVEEM LRYVESKPTN ELSQRLQRFV TKRPMPVSDS FVCLRPVDFQ RLTQVIEQRR RVLQRQ REE YHGVYEHLAG LITSIDIHDL DASDLNRREI LKALQPLDDN AKQELFRLGN AKMLELQMDL DRLSTQLLTR VHNHILN GF LPVEDLKQME RVVEQVLRLF YDLRDLKLCD GSYEEGFVVI REQLSYLMTG TVRDNVPLLQ EILQLRHAYQ QATQQNEG R LTQIHDLLHV IETLVRDPGS RGSALTLALV QEQLAQLEAL GGLQLPEVQQ RLQNAQLALS RLYEEEEETQ RFLDGLSYD DPPNEQTIKR HPQLREMLRR DEQTRLRLIN AVLSMFHTLV MRLARDESPR PTFFDAVSLL LQQLPPDSHE REDLRAANAT YAQMVKKLE QIEKAGTGAS EKRFQALREL VYFFRNHEYF FQHMVGRLGV GPQVTELYER YQHEMEEQHL ERLEREWQEE A GKLTVTSV EDVQRVLARA PSHRVMHQMQ QTLTTKMQDF LDKEKRKQEE QQRQLLDGYQ KKVQQDLQRV VDAVKGEMLS TI PHQPLEA TLELLLGLDQ RAQPLLDKFN QDLLSALQQL SKKLDGRINE CLHGVLTGDV ERRCHPHREA AMQTQASLNH LDQ ILGPQL LIHETQQALQ HAVHQAQFIE KCQQGDPTTA ITGSEFEGDF ARYRSSQQKM EEQLQETRQQ MTETSERLDR SLRQ DPGSS SVTRVPEKPF KGQELAGRIT PPPADFQQPV FKTLLDQQAD AARKALSDEA DLLNQKVQTQ LRQRDEQLST AQNLW TDLV TRHKMSGGLD VTTPDAKALM EKPLETLREL LGKATQQLPY LSAERTVRWM LAFLEEALAQ ITADPTHPHH GSRTHY RNL QQQAVESAVT LAHQIEQNAA CENFIAQHQE ATANGASTPR VDMVQAVEAV WQRLEPGRVA GGAARHQKVQ ELLQRLG QT LGDLELQETL ATEYFALLHG IQTFSYGLDF RSQLEKIRDL RTRFAELAKR RGTRLSNEGV LPNPRKPQAT TSLGAFTR G LNALERHVQL GHQYLLNKLN GSSLVYRLED IPSVLPATHE TDPALIMRDR LRRLCFARHH DTFLEVVDVF GMRQIVTQA GEPIHLVTDY GNVAFKYLAL RDDGRPLAWR RRCSGGGLKN VVTTRYKAIT VAVAVCQTLR TFWPQISQYD LRPYLTQHQS HTHPAETHT LHNLKLFCYL VSTAWHQRID TQQELTAADR VGSGEGGDVG EQRPGRGTVL RLSLQEFCVL IAALYPEYIY T VLKYPVQM SLPSLTAHLH QDVIHAVVNN THKMPPDHLP EQVKAFCITP TQWPAMQLNK LFWENKLVQQ LCQVGPQKST PP LGKLWLY AMATLVFPQD MLQCLWLELK PQYAETYASV SELVQTLFQI FTQQCEMVTE GYTQPQLPTG EPVLQMIRVP RQD TTTTDT NTTTEPGLLD VFIQTETALD YALGSWLFGI PVCLGVHVAD LLKGQRILVA RHLEYTSRDR DFLRIQRSRD LNLS QLLQD TWTETPLEHC WLQAQIRRLR DYLRFPTRLE FIPLVIYNAQ DHTVVRVLRP PSTFEQDHSR LVLDEAFPTF PLYDQ DDNS SADNIAASGA APTPPVPFNR VPVNIQFLRE NPPPIARVQQ PPRRHRHRAA AAADDDGQID HVQDDTSRTA DSALVS TAF GGSVFQENRL GETPLCRDEL VAVAPGAAST SFASPPITVL TQNVLSALEI LRLVRLDLRQ LAQSVQDTIQ HMRFLYL L UniProtKB: Large tegument protein deneddylase |
-Macromolecule #5: Capsid vertex component 2
| Macromolecule | Name: Capsid vertex component 2 / type: protein_or_peptide / ID: 5 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 71.26957 KDa |
| Sequence | String: MSLLHTFWRL PVAVFFEPHE ENVLRCPERV LRRLLEDAAV TMRGGGWRED VLMDRVRKRY LRQELRDLGH RVQTYCEDLE GRVSEAEAL LNQQCELDEG PSPRTLLQPP CRPRSSSPGT GVAGASAVPH GLYSRHDAIT GPAAAPSDVV APSDAVAASA A AGASSTWL ...String: MSLLHTFWRL PVAVFFEPHE ENVLRCPERV LRRLLEDAAV TMRGGGWRED VLMDRVRKRY LRQELRDLGH RVQTYCEDLE GRVSEAEAL LNQQCELDEG PSPRTLLQPP CRPRSSSPGT GVAGASAVPH GLYSRHDAIT GPAAAPSDVV APSDAVAASA A AGASSTWL AQCAERPLPG NVPSYFGITQ NDPFIRFHTD FRGEVVNTMF ENASTWTFSF GIWYYRLKRG LYTQPRWKRV YH LAQMDNF SISQELLLGV VNALENVTVY PTYDCVLSDL EAAACLLAAY GHALWEGRDP PDSVATVLGE LPQLLPRLAD DVS REIAAW EGPVAAGNNY YAYRDSPDLR YYMPLSGGRH YHPGTFDRHV LVRLFHKRGV IQHLPGYGTI TEELVQERLS GQVR DDVLS LWSRRLLVGK LGRDVPVFVH EQQYLRSGLT CLAGLLLLWK VTNADSVFAP RTGKFTLADL LGSDAVAGGG LPGGR AGGE EEGYGGRHGR VRNFEFLVRY YIGPWYARDP AVTLSQLFPG LALLAVTESV RSGWDPSRRE DSAGGGDGGG AVLMQL SKS NPVADYMFAQ SSKQYGDLRR LEVHDALLFH YEHGLGRLLS VTLPRHRVST LGSSLFNVND IYELLYFLVL GFLPSVA VL UniProtKB: Capsid vertex component 2 |
-Macromolecule #6: Capsid vertex component 1
| Macromolecule | Name: Capsid vertex component 1 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 68.567211 KDa |
| Sequence | String: METHLYSDLA FEARFADDEQ LPLHLVLDQE VLSNEEAETL RYVYYRNVDS AGRSTGRAPG GDEDDAPASD DAEDAVGGDR AFDRERRTW QRACFRVLPR PLELLDYLRQ SGLTVTLEKE QRVRMFYAVF TTLGLRCPDN RLSGAQTLHL RLVWPDGSYR D WEFLARDL ...String: METHLYSDLA FEARFADDEQ LPLHLVLDQE VLSNEEAETL RYVYYRNVDS AGRSTGRAPG GDEDDAPASD DAEDAVGGDR AFDRERRTW QRACFRVLPR PLELLDYLRQ SGLTVTLEKE QRVRMFYAVF TTLGLRCPDN RLSGAQTLHL RLVWPDGSYR D WEFLARDL LREEMEANKR DRQHQLATTT NHRRRGGLRN NLDNGSDRRL PEAAVASLET AVSTPFFEIP NGAGTSSANG DG RFSNLEQ RVARLLRGDE EFIYHAGPLE PPSKIRGHEL VQLRLDVNPD LMYATDPHDR DEVARTDEWK GAGVSRLREV WDV QHRVRL RVLWYVNSFW RSRELSYDDH EVELYRALDA YRARIAVEYV LIRAVRDEIY AVLRRDGGAL PQRFACHVSR NMSW RVVWE LCRHALALWM DWADVRSCII KALTPRLSRG AAAAAQRARR QRERSAPKPQ ELLFGPRNES GPPAEQTWYA DVVRC VRAQ VDLGVEVRAA RCPRTGLWIV RDRRGRLRRW LSQPEVCVLY VTPDLDFYWV LPGGFAVSSR VTLHGLAQRA LRDRFQ NFE AVLARGMHVE AGRQEPETPR VSGRRLPFDD L UniProtKB: Capsid vertex component 1 |
-Macromolecule #7: Triplex capsid protein 1
| Macromolecule | Name: Triplex capsid protein 1 / type: protein_or_peptide / ID: 7 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 33.07127 KDa |
| Sequence | String: MDARAVAKRP RDPADEDNEL VTALKAKREV NTISVRYLYH ADHQALTARF FVPEGLVEFE AQPGALLIRM ETGCDSPRHL YISLYLLGI RASNVSASTR CLLESVYTAS AARAALQWLD LGPHLLHRRL ETLGCVKTVS LGITSLLTCV MRGYLYNTLK T EVFALMIP ...String: MDARAVAKRP RDPADEDNEL VTALKAKREV NTISVRYLYH ADHQALTARF FVPEGLVEFE AQPGALLIRM ETGCDSPRHL YISLYLLGI RASNVSASTR CLLESVYTAS AARAALQWLD LGPHLLHRRL ETLGCVKTVS LGITSLLTCV MRGYLYNTLK T EVFALMIP KDMYLTWEET RGRLQYVYLI IVYDYDGPET RPGIYVLTSS IAHWQTLVDV ARGKFARERC SFVNRRITRP RQ IPLCTGV IQKLGWCLAD DIHTSFLVHK ELKLSVVRLD NFSVELGDFR EFV UniProtKB: Triplex capsid protein 1 |
-Macromolecule #8: Triplex capsid protein 2
| Macromolecule | Name: Triplex capsid protein 2 / type: protein_or_peptide / ID: 8 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human herpesvirus 5 strain AD169 Human herpesvirus 5 strain AD169 |
| Molecular weight | Theoretical: 34.63575 KDa |
| Sequence | String: MAAMEANIFC TFDHKLSIAD VGKLTKLVAA VVPIPQRLHL IKHYQLGLHQ FVDHTRGYVR LRGLLRNMTL TLMRRVEGNQ ILLHVPTHG LLYTVLNTGP VTWEKGDALC VLPPLFHGPL ARENLLTLGQ WELVLPWIVP MPLALEINQR LLIMGLFSLD R SYEEVKAA ...String: MAAMEANIFC TFDHKLSIAD VGKLTKLVAA VVPIPQRLHL IKHYQLGLHQ FVDHTRGYVR LRGLLRNMTL TLMRRVEGNQ ILLHVPTHG LLYTVLNTGP VTWEKGDALC VLPPLFHGPL ARENLLTLGQ WELVLPWIVP MPLALEINQR LLIMGLFSLD R SYEEVKAA VQQLQTITFR DATFTIPDPV IDQHLLIDMK TACLSMSMVA NLASELTMTY VRKLALEDSS MLLVKCQELL MR LDRERSV GEPRTPARPQ HVSPDDEIAR LSALFVMLRQ LDDLIREQVV FTVCDVSPDN KSATCIFKG UniProtKB: Triplex capsid protein 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average exposure time: 8.0 sec. / Average electron dose: 47.2 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)