[English] 日本語
 Yorodumi
Yorodumi- EMDB-41125: Zophobas morio black wasting virus strain NJ2-molitor virion structure -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Zophobas morio black wasting virus strain NJ2-molitor virion structure | |||||||||
 Map data Map data | Main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Capsid / Virion / Parvovirus / Densovirus / Invertebrate / Insect / Pathogen / ssDNA / VIRUS / VIRUS-DNA complex | |||||||||
| Biological species |  Zophobas morio black wasting virus / Zophobas morio black wasting virus /  Zophobas morio densovirus Zophobas morio densovirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | |||||||||
 Authors Authors | Penzes JJ / Kaelber JT | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Sequencing-free discovery by cryo-EM of a pathogenic parvovirus causing mass mortality of farmed beetles Authors: Penzes JJ / Holm M / Firlar E / Kaelber JT | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41125.map.gz emd_41125.map.gz | 2.6 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41125-v30.xml emd-41125-v30.xml emd-41125.xml emd-41125.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_41125.png emd_41125.png | 82 KB | ||
| Filedesc metadata |  emd-41125.cif.gz emd-41125.cif.gz | 6.2 KB | ||
| Others |  emd_41125_half_map_1.map.gz emd_41125_half_map_1.map.gz emd_41125_half_map_2.map.gz emd_41125_half_map_2.map.gz | 2.5 GB 2.5 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41125 http://ftp.pdbj.org/pub/emdb/structures/EMD-41125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41125 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41125 | HTTPS FTP |
-Validation report
| Summary document |  emd_41125_validation.pdf.gz emd_41125_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41125_full_validation.pdf.gz emd_41125_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_41125_validation.xml.gz emd_41125_validation.xml.gz | 27.7 KB | Display | |
| Data in CIF |  emd_41125_validation.cif.gz emd_41125_validation.cif.gz | 33.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41125 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41125 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41125 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41125 | HTTPS FTP |
-Related structure data
| Related structure data |  8t9xMC  8t9cC  8t9eC  8ta7C  8tjeC M: atomic model generated by this map C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_41125.map.gz / Format: CCP4 / Size: 2.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41125.map.gz / Format: CCP4 / Size: 2.7 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.63 Å | ||||||||||||||||||||||||||||||||||||
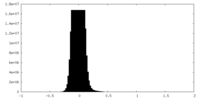
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map of D 1000275531 em-volume P1.map.V3
| File | emd_41125_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of D_1000275531_em-volume_P1.map.V3 | ||||||||||||
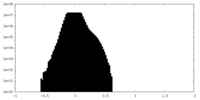
| Projections & Slices |
| ||||||||||||
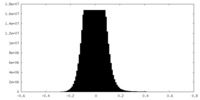
| Density Histograms |
-Half map: Half map of D 1000275531 em-volume P1.map.V3
| File | emd_41125_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map of D_1000275531_em-volume_P1.map.V3 | ||||||||||||
| Projections & Slices |
| ||||||||||||
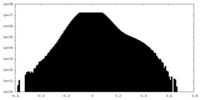
| Density Histograms |
- Sample components
Sample components
-Entire : Zophobas morio densovirus
| Entire | Name:  Zophobas morio densovirus Zophobas morio densovirus |
|---|---|
| Components |
|
-Supramolecule #1: Zophobas morio densovirus
| Supramolecule | Name: Zophobas morio densovirus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: Purified from T. molitor larvae, which were asymptomatic NCBI-ID: 2750924 / Sci species name: Zophobas morio densovirus / Sci species strain: NJ2-molitor / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Tenebrio molitor (yellow mealworm) Tenebrio molitor (yellow mealworm) |
| Virus shell | Shell ID: 1 / Diameter: 28.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| Molecular weight | Theoretical: 48.085629 KDa |
| Sequence | String: ATAILRPIGL HVEKFQQTYR KKWRFLTSAN ANVILAEAAS GERPARWALT TGMASIPWEY LFFYMSPAEY NRMKNYPGTF AKSASVRIR TWNTRVAFQT GDTQTANATL NQNKFLQVAK GIRSIPFICS TNRKYTYSDT EPMQPTGFAT LTSYEYRDGL K IAMYGYDN ...String: ATAILRPIGL HVEKFQQTYR KKWRFLTSAN ANVILAEAAS GERPARWALT TGMASIPWEY LFFYMSPAEY NRMKNYPGTF AKSASVRIR TWNTRVAFQT GDTQTANATL NQNKFLQVAK GIRSIPFICS TNRKYTYSDT EPMQPTGFAT LTSYEYRDGL K IAMYGYDN DSADFAKKPP ADATGAEIYL QDYLTIYTND ARATTGTKIL AGFPPYKNFI EEFDASACIN TDVVAMDYDF SY APLVPQF APVPNNLITQ NYNASYPAGT KNEVTAVKTT DSSQATPPTQ VRNAPRKYIQ GPNADTTFFD EEQNYLRVPI EQG GIFEEV NVETVHDTQM PSINVGIRAV PKLTTIDETT QANSWLDAQG YFEVDCVLTT ESVDPYTYIK GGCYSANTKS QLQY FASDG RPIAKVYDNP NVYGRMQMIK TVKP |
-Macromolecule #2: DNA (5'-D(P*CP*GP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*GP*A)-3') / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| Molecular weight | Theoretical: 886.637 Da |
| Sequence | String: (DC)(DG)(DA) |
-Macromolecule #3: DNA (5'-D(P*CP*AP*GP*GP*CP*CP*AP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*CP*AP*GP*GP*CP*CP*AP*AP*A)-3') / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| Molecular weight | Theoretical: 2.733827 KDa |
| Sequence | String: (DC)(DA)(DG)(DG)(DC)(DC)(DA)(DA)(DA) |
-Macromolecule #4: DNA (5'-D(P*TP*CP*GP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(P*TP*CP*GP*AP*A)-3') / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Zophobas morio black wasting virus Zophobas morio black wasting virus |
| Molecular weight | Theoretical: 1.504037 KDa |
| Sequence | String: (DT)(DC)(DG)(DA)(DA) |
-Macromolecule #5: water
| Macromolecule | Name: water / type: ligand / ID: 5 / Number of copies: 80 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Component - Concentration: 1.0 x / Component - Formula: PBS / Component - Name: Phosphate-buffered saline |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 300 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | Purified virus from homogenized T. molitor larval tissue |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average exposure time: 3.0 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.2 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8t9x: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)