+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli SIR2-HerA complex (dodecamer SIR2 pentamer HerA) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HerA / SIR2 / NADase / ATPase / Anti-phage system / IMMUNE SYSTEM | |||||||||
| Function / homology | Helicase HerA, central domain / Helicase HerA, central domain / SIR2-like domain / SIR2-like domain / hydrolase activity / P-loop containing nucleoside triphosphate hydrolase / SIR2-like domain-containing protein / Nucleoside triphosphate hydrolase Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.89 Å | |||||||||
 Authors Authors | Shen ZF / Lin QP / Fu TM | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2023 Journal: Mol Cell / Year: 2023Title: Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense. Authors: Zhangfei Shen / Qingpeng Lin / Xiao-Yuan Yang / Elizabeth Fosuah / Tian-Min Fu /  Abstract: SIR2-HerA, a bacterial two-protein anti-phage defense system, induces bacterial death by depleting NAD upon phage infection. Biochemical reconstitution of SIR2, HerA, and the SIR2-HerA complex ...SIR2-HerA, a bacterial two-protein anti-phage defense system, induces bacterial death by depleting NAD upon phage infection. Biochemical reconstitution of SIR2, HerA, and the SIR2-HerA complex reveals a dynamic assembly process. Unlike other ATPases, HerA can form various oligomers, ranging from dimers to nonamers. When assembled with SIR2, HerA forms a hexamer and converts SIR2 from a nuclease to an NAD hydrolase, representing an unexpected regulatory mechanism mediated by protein assembly. Furthermore, high concentrations of ATP can inhibit NAD hydrolysis by the SIR2-HerA complex. Cryo-EM structures of the SIR2-HerA complex reveal a giant supramolecular assembly up to 1 MDa, with SIR2 as a dodecamer and HerA as a hexamer, crucial for anti-phage defense. Unexpectedly, the HerA hexamer resembles a spiral staircase and exhibits helicase activities toward dual-forked DNA. Together, we reveal the supramolecular assembly of SIR2-HerA as a unique mechanism for switching enzymatic activities and bolstering anti-phage defense strategies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40763.map.gz emd_40763.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40763-v30.xml emd-40763-v30.xml emd-40763.xml emd-40763.xml | 18.6 KB 18.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40763_fsc.xml emd_40763_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_40763.png emd_40763.png | 75.2 KB | ||
| Filedesc metadata |  emd-40763.cif.gz emd-40763.cif.gz | 6.1 KB | ||
| Others |  emd_40763_additional_1.map.gz emd_40763_additional_1.map.gz emd_40763_half_map_1.map.gz emd_40763_half_map_1.map.gz emd_40763_half_map_2.map.gz emd_40763_half_map_2.map.gz | 107.9 MB 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40763 http://ftp.pdbj.org/pub/emdb/structures/EMD-40763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40763 | HTTPS FTP |
-Validation report
| Summary document |  emd_40763_validation.pdf.gz emd_40763_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40763_full_validation.pdf.gz emd_40763_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40763_validation.xml.gz emd_40763_validation.xml.gz | 19.2 KB | Display | |
| Data in CIF |  emd_40763_validation.cif.gz emd_40763_validation.cif.gz | 24.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40763 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40763 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40763 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40763 | HTTPS FTP |
-Related structure data
| Related structure data |  8subMC  8su9C  8suwC  8sxxC  8uaeC  8uafC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40763.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40763.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||
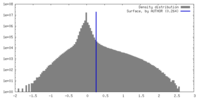
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_40763_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
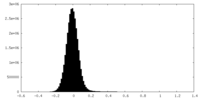
| Density Histograms |
-Half map: #2
| File | emd_40763_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40763_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SIR2-HerA complex 2(12:5)
| Entire | Name: SIR2-HerA complex 2(12:5) |
|---|---|
| Components |
|
-Supramolecule #1: SIR2-HerA complex 2(12:5)
| Supramolecule | Name: SIR2-HerA complex 2(12:5) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: pentamer HerA and dodecamer Sir2 form a complex |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: SIR2-like domain-containing protein
| Macromolecule | Name: SIR2-like domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.817664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIYQGGNKL NEDDFRSHVY SLCQLDNVGV LLGAGASVGC GGKTMKDVWK SFKQNYPELL GALIDKYLLV SQIDSDNNLV NVELLIDEA TKFLSVAKTR RCEDEEEEFR KILSSLYKEV TKAALLTGEQ FREKNQGKKD AFKYHKELIS KLISNRQPGQ S APAIFTTN ...String: MSIYQGGNKL NEDDFRSHVY SLCQLDNVGV LLGAGASVGC GGKTMKDVWK SFKQNYPELL GALIDKYLLV SQIDSDNNLV NVELLIDEA TKFLSVAKTR RCEDEEEEFR KILSSLYKEV TKAALLTGEQ FREKNQGKKD AFKYHKELIS KLISNRQPGQ S APAIFTTN YDLALEWAAE DLGIQLFNGF SGLHTRQFYP QNFDLAFRNV NAKGEARFGH YHAYLYKLHG SLTWYQNDSL TV NEVSASQ AYDEYINDII NKDDFYRGQH LIYPGANKYS HTIGFVYGEM FRRFGEFISK PQTALFINGF GFGDYHINRI ILG ALLNPS FHVVIYYPEL KEAITKVSKG GGSEAEKAIV TLKNMAFNQV TVVGGGSKAY FNSFVEHLPY PVLFPRDNIV DELV EAIAN LSKGEGNVPF UniProtKB: SIR2-like domain-containing protein |
-Macromolecule #2: Nucleoside triphosphate hydrolase
| Macromolecule | Name: Nucleoside triphosphate hydrolase / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.431992 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLFKLTEIS AIGYVVGLEG ERIRINLHEG LQGRLASHRK GVSSVTQPGD LIGFDAGNIL VVARVTDMAF VEADKAHKAN VGTSDLADI PLRQIIAYAI GFVKRELNGY VFISEDWRLP ALGSSAVPLT SDFLNIIYSI DKEELPKAVE LGVDSRTKTV K IFASVDKL ...String: MSLFKLTEIS AIGYVVGLEG ERIRINLHEG LQGRLASHRK GVSSVTQPGD LIGFDAGNIL VVARVTDMAF VEADKAHKAN VGTSDLADI PLRQIIAYAI GFVKRELNGY VFISEDWRLP ALGSSAVPLT SDFLNIIYSI DKEELPKAVE LGVDSRTKTV K IFASVDKL LSRHLAVLGS TGYGKSNFNA LLTRKVSEKY PNSRIVIFDI NGEYAQAFTG IPNVKHTILG ESPNVDSLEK KQ QKGELYS EEYYCYKKIP YQALGFAGLI KLLRPSDKTQ LPALRNALSA INRTHFKSRN IYLEKDDGET FLLYDDCRDT NQS KLAEWL DLLRRRRLKR TNVWPPFKSL ATLVAEFGCV AADRSNGSKR DAFGFSNVLP LVKIIQQLAE DIRFKSIVNL NGGG ELADG GTHWDKAMSD EVDYFFGKEK GQENDWNVHI VNMKNLAQDH APMLLSALLE MFAEILFRRG QERSYPTVLL LEEAH HYLR DPYAEIDSQI KAYERLAKEG RKFKCSLIVS TQRPSELSPT VLAMCSNWFS LRLTNERDLQ ALRYAMESGN EQILKQ ISG LPRGDAVAFG SAFNLPVRIS INQARPGPKS SDAVFSEEWA NCTELRC UniProtKB: Nucleoside triphosphate hydrolase |
-Macromolecule #3: [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]ME...
| Macromolecule | Name: [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE type: ligand / ID: 3 / Number of copies: 12 / Formula: AR6 |
|---|---|
| Molecular weight | Theoretical: 559.316 Da |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 5 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)