[English] 日本語
 Yorodumi
Yorodumi- EMDB-40473: Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | |||||||||
 Map data Map data | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Multienzyme complex / carboxylase / LIGASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationpropionyl-CoA carboxylase / urea carboxylase activity / methylcrotonoyl-CoA carboxylase activity / propionyl-CoA carboxylase activity / acetyl-CoA carboxylase complex / lipid catabolic process / mitochondrion / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 8.62 Å | |||||||||
 Authors Authors | Lee JKJ / Liu YT / Hu JJ / Aphasizheva I / Aphasizhev R / Zhou ZH | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Struct Biol X / Year: 2023 Journal: J Struct Biol X / Year: 2023Title: CryoEM reveals oligomeric isomers of a multienzyme complex and assembly mechanics. Authors: Jane K J Lee / Yun-Tao Liu / Jason J Hu / Inna Aphasizheva / Ruslan Aphasizhev / Z Hong Zhou /  Abstract: Propionyl-CoA carboxylase (PCC) is a multienzyme complex consisting of up to six α-subunits and six β-subunits. Belonging to a metabolic pathway converging on the citric acid cycle, it is present ...Propionyl-CoA carboxylase (PCC) is a multienzyme complex consisting of up to six α-subunits and six β-subunits. Belonging to a metabolic pathway converging on the citric acid cycle, it is present in most forms of life and irregularities in its assembly lead to serious illness in humans, known as propionic acidemia. Here, we report the cryogenic electron microscopy (cryoEM) structures and assembly of different oligomeric isomers of endogenous PCC from the parasitic protozoan (LtPCC). These structures and their statistical distribution reveal the mechanics of PCC assembly and disassembly at equilibrium. We show that, in solution, endogenous LtPCC β-subunits form stable homohexamers, to which different numbers of α-subunits attach. Sorting LtPCC particles into seven classes (i.e., oligomeric formulae αβ, αβ, αβ, αβ, αβ, αβ, αβ) enables formulation of a model for PCC assembly. Our results suggest how multimerization regulates PCC enzymatic activity and showcase the utility of cryoEM in revealing the statistical mechanics of reaction pathways. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40473.map.gz emd_40473.map.gz | 201.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40473-v30.xml emd-40473-v30.xml emd-40473.xml emd-40473.xml | 15.8 KB 15.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_40473.png emd_40473.png | 77 KB | ||
| Filedesc metadata |  emd-40473.cif.gz emd-40473.cif.gz | 5.9 KB | ||
| Others |  emd_40473_half_map_1.map.gz emd_40473_half_map_1.map.gz emd_40473_half_map_2.map.gz emd_40473_half_map_2.map.gz | 171.4 MB 171.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40473 http://ftp.pdbj.org/pub/emdb/structures/EMD-40473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40473 | HTTPS FTP |
-Validation report
| Summary document |  emd_40473_validation.pdf.gz emd_40473_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40473_full_validation.pdf.gz emd_40473_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40473_validation.xml.gz emd_40473_validation.xml.gz | 15.3 KB | Display | |
| Data in CIF |  emd_40473_validation.cif.gz emd_40473_validation.cif.gz | 18.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40473 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40473 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40473 | HTTPS FTP |
-Related structure data
| Related structure data |  8sgyMC  8sgxC  8sgzC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40473.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Leishmania tarentolae propionyl-CoA carboxylase (alpha-5-beta-6) | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40473_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
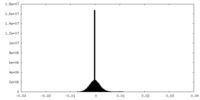
| Projections & Slices |
| ||||||||||||
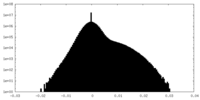
| Density Histograms |
-Half map: Half Map 2
| File | emd_40473_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
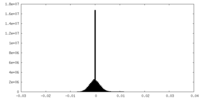
| Projections & Slices |
| ||||||||||||
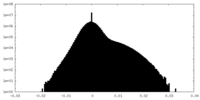
| Density Histograms |
- Sample components
Sample components
-Entire : Leishmania tarentolae propionyl-CoA carboxylase
| Entire | Name: Leishmania tarentolae propionyl-CoA carboxylase |
|---|---|
| Components |
|
-Supramolecule #1: Leishmania tarentolae propionyl-CoA carboxylase
| Supramolecule | Name: Leishmania tarentolae propionyl-CoA carboxylase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) |
-Macromolecule #1: propionyl-CoA carboxylase
| Macromolecule | Name: propionyl-CoA carboxylase / type: protein_or_peptide / ID: 1 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) |
| Molecular weight | Theoretical: 73.277656 KDa |
| Sequence | String: VLVANRGEIA CRVMATCRRL GIKTVAVYST ADEQAKHVKV ADESVCIGPP ASVESYLCID KIVDACKKTG AQAVHPGYGF LSENGEFQS ALQKNNIVFV GPDAHSIESM GDKIESKRLA QRAGVTCIPG FIGEVKTHED LLRFAREIGY PVMIKASGGG G GKGMRVAY ...String: VLVANRGEIA CRVMATCRRL GIKTVAVYST ADEQAKHVKV ADESVCIGPP ASVESYLCID KIVDACKKTG AQAVHPGYGF LSENGEFQS ALQKNNIVFV GPDAHSIESM GDKIESKRLA QRAGVTCIPG FIGEVKTHED LLRFAREIGY PVMIKASGGG G GKGMRVAY NDTQCVEYYD MCREEAKAAF HSDKMLVERF IDHPRHIEIQ VIADRRGNTV YLPERECSIQ RRNQKVIEEA PS VLLDATT RKAMGEEAVA MARAVQYVSA GTVENVVNPQ KQFYFLEMNT RLQVEHPITE EITGVDLVEQ MLRAAADLPL SIT QDDITI NGHATECRVY AEDPMKNYFP SIGRLTMYQE PTGAGVRCDS GIIEGSQISV YYDPLICKLS TWGRDRAECI GRME KALDE YVIRGLRHNI CLLRDVVTEP RYRSGSITTN YLQEQYPNGF KKAELTAEEM QLMYEVAACV HLKRERLHYT QGTAP SERQ LYLSVGAGQE GETPVYVRYL DDSHFEIGAS KHGPFRKMEV VWKASYPIIR VKDGEAETVL QFWGTNEVTY GMQMRG TTF DVNVMSDLQS TLAHFVPITE ATTNTKQILS PMPGVIVAIK VQPGQMVVAG EELLTLEAMK MRNKIHAQAD GKVKEVK VK LGATVEDNEV LVELE UniProtKB: propionyl-CoA carboxylase |
-Macromolecule #2: Propionyl-coa carboxylase beta chain, putative
| Macromolecule | Name: Propionyl-coa carboxylase beta chain, putative / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) |
| Molecular weight | Theoretical: 53.587285 KDa |
| Sequence | String: PTAAEDLRHK KKRLTAMERV QLFCDPGTFR ERDALVEHEC HNFGMEKRKV PGDGFITGTG KVFGRPVFLF SHDFTVFGGS LSRTNAAKV VRIMEEAAKI GVPVIGFNDS GGARIHEGVD SLAGYADIFL RNTLFSGVIP QISVIMGPCA GGAVYSPAIT D FTFMVETS ...String: PTAAEDLRHK KKRLTAMERV QLFCDPGTFR ERDALVEHEC HNFGMEKRKV PGDGFITGTG KVFGRPVFLF SHDFTVFGGS LSRTNAAKV VRIMEEAAKI GVPVIGFNDS GGARIHEGVD SLAGYADIFL RNTLFSGVIP QISVIMGPCA GGAVYSPAIT D FTFMVETS SYMFVTGPEV VSAVGGKLVT KDELGGPHVH ATKSGVSAGT FPNDIVAMAQ LRRLYSYLPL SNRDPVPVLP TA DERYRDV SSLNTVVPTE VKEAYDMRDV IYPVIDHDSF FEIQPQFAKN IICGFARVEG RSVCIIANQP KVQAGVLDID SSV KGARMV RFADAFNIPI ITFVDVPGFL PGVQQEYGGI IRHGAKLLYA YAEATVPKVT IITRKAYGGA YDVMSSKHLR GDSN YAWPH AEIAVMGAAG ACKLLYSKET AEQQAQRIAD YEKTFCTPLS AARKGFVDAV IDPSETRMRV CEDLERLARK QLQNP WKKH GNIPL UniProtKB: Propionyl-coa carboxylase beta chain, putative |
-Macromolecule #3: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL
| Macromolecule | Name: 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL type: ligand / ID: 3 / Number of copies: 6 / Formula: BTI |
|---|---|
| Molecular weight | Theoretical: 228.311 Da |
| Chemical component information |  ChemComp-BTI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 8.62 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 16034 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X