[English] 日本語
 Yorodumi
Yorodumi- EMDB-40308: Sub-tomogram average of the RSV F pairs from the surface of nativ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Sub-tomogram average of the RSV F pairs from the surface of native virions released from RSV-infected BEAS-2B cells cultured on EM grids collected via montage parallel array cryo-tomography (MPACT) | |||||||||
 Map data Map data | STA of RSV glycoprotein F pair on the surface of native RSV virion collected via parallel array montage cryo-electron tomography | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | natively folded / class I viral fusion protein / pre-fusion native state / whole virion / VIRUS | |||||||||
| Biological species |  Respiratory syncytial virus A2 Respiratory syncytial virus A2 | |||||||||
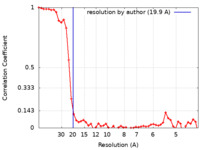
| Method | subtomogram averaging / cryo EM / Resolution: 19.9 Å | |||||||||
 Authors Authors | Yang J / Wright ER | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Methods / Year: 2023 Journal: Nat Methods / Year: 2023Title: Correlative montage parallel array cryo-tomography for in situ structural cell biology. Authors: Jie E Yang / Matthew R Larson / Bryan S Sibert / Joseph Y Kim / Daniel Parrell / Juan C Sanchez / Victoria Pappas / Anil Kumar / Kai Cai / Keith Thompson / Elizabeth R Wright /  Abstract: Imaging large fields of view while preserving high-resolution structural information remains a challenge in low-dose cryo-electron tomography. Here we present robust tools for montage parallel array ...Imaging large fields of view while preserving high-resolution structural information remains a challenge in low-dose cryo-electron tomography. Here we present robust tools for montage parallel array cryo-tomography (MPACT) tailored for vitrified specimens. The combination of correlative cryo-fluorescence microscopy, focused-ion-beam milling, substrate micropatterning, and MPACT supports studies that contextually define the three-dimensional architecture of cells. To further extend the flexibility of MPACT, tilt series may be processed in their entirety or as individual tiles suitable for sub-tomogram averaging, enabling efficient data processing and analysis. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40308.map.gz emd_40308.map.gz | 7.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40308-v30.xml emd-40308-v30.xml emd-40308.xml emd-40308.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40308_fsc.xml emd_40308_fsc.xml | 6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40308.png emd_40308.png | 28.1 KB | ||
| Filedesc metadata |  emd-40308.cif.gz emd-40308.cif.gz | 5.2 KB | ||
| Others |  emd_40308_half_map_1.map.gz emd_40308_half_map_1.map.gz emd_40308_half_map_2.map.gz emd_40308_half_map_2.map.gz | 7.3 MB 7.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40308 http://ftp.pdbj.org/pub/emdb/structures/EMD-40308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40308 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40308 | HTTPS FTP |
-Validation report
| Summary document |  emd_40308_validation.pdf.gz emd_40308_validation.pdf.gz | 605.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40308_full_validation.pdf.gz emd_40308_full_validation.pdf.gz | 605.2 KB | Display | |
| Data in XML |  emd_40308_validation.xml.gz emd_40308_validation.xml.gz | 11.1 KB | Display | |
| Data in CIF |  emd_40308_validation.cif.gz emd_40308_validation.cif.gz | 13.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40308 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40308 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40308 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40308 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40308.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40308.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | STA of RSV glycoprotein F pair on the surface of native RSV virion collected via parallel array montage cryo-electron tomography | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.503 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: STA of RSV glycoprotein F pair on the...
| File | emd_40308_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | STA of RSV glycoprotein F pair on the surface of native RSV virion collected via parallel array montage cryo-electron tomography | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: STA of RSV glycoprotein F pair on the...
| File | emd_40308_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | STA of RSV glycoprotein F pair on the surface of native RSV virion collected via parallel array montage cryo-electron tomography | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Respiratory syncytial virus A2
| Entire | Name:  Respiratory syncytial virus A2 Respiratory syncytial virus A2 |
|---|---|
| Components |
|
-Supramolecule #1: Respiratory syncytial virus A2
| Supramolecule | Name: Respiratory syncytial virus A2 / type: virus / ID: 1 / Parent: 0 Details: The strain was kindly provided by Dr. Martin L. Moore. The reference is PMCID: PMC3492879/PMID:23062737 NCBI-ID: 1972429 / Sci species name: Respiratory syncytial virus A2 / Sci species strain: rA2-mK+ / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 17 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 303 K / Instrument: GATAN CRYOPLUNGE 3 |
| Details | Respiratory syncytial virus released from infected whole BEAS-2B mammalian cells (human non-tumorigenic lung epithelial cell) |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Average exposure time: 0.5 sec. / Average electron dose: 1.8 e/Å2 Details: Tilt series of defocus collection ranging from -3 to -6 micrometer, with a step of -0.5 micrometer |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 6.8500000000000005 µm / Calibrated defocus min: 3.3200000000000003 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 6.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)