+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | |||||||||
 Map data Map data | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | efflux pump / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationxenobiotic transport / efflux transmembrane transporter activity / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
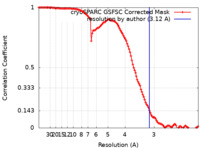
| Method | single particle reconstruction / cryo EM / Resolution: 3.12 Å | |||||||||
 Authors Authors | Zhang Z | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Microbiol Spectr / Year: 2023 Journal: Microbiol Spectr / Year: 2023Title: Cryo-Electron Microscopy Structures of a Campylobacter Multidrug Efflux Pump Reveal a Novel Mechanism of Drug Recognition and Resistance. Authors: Zhemin Zhang / Nicholas Lizer / Zuowei Wu / Christopher E Morgan / Yuqi Yan / Qijing Zhang / Edward W Yu /  Abstract: Campylobacter jejuni is a bacterium that is commonly present in the intestinal tracts of animals. It is also a major foodborne pathogen that causes gastroenteritis in humans. The most predominant and ...Campylobacter jejuni is a bacterium that is commonly present in the intestinal tracts of animals. It is also a major foodborne pathogen that causes gastroenteritis in humans. The most predominant and clinically important multidrug efflux system in C. jejuni is the CmeABC (Campylobacter multidrug efflux) pump, a tripartite system that includes an inner membrane transporter (CmeB), a periplasmic fusion protein (CmeA), and an outer membrane channel protein (CmeC). This efflux protein machinery mediates resistance to a number of structurally diverse antimicrobial agents. A recently identified CmeB variant, termed resistance enhancing CmeB (RE-CmeB), can increase its multidrug efflux pump activity, likely by influencing antimicrobial recognition and extrusion. Here, we report structures of RE-CmeB in its apo form as well as in the presence of four different drugs by using single-particle cryo-electron microscopy (cryo-EM). Coupled with mutagenesis and functional studies, this structural information allows us to identify critical amino acids that are important for drug resistance. We also report that RE-CmeB utilizes a somewhat unique subset of residues to bind different drugs, thereby optimizing its ability to accommodate different compounds with distinct scaffolds. These findings provide insights into the structure-function relationship of this newly emerged antibiotic efflux transporter variant in Campylobacter. Campylobacter jejuni has emerged as one of the most problematic and highly antibiotic-resistant pathogens, worldwide. The Centers for Disease Control and Prevention have designated antibiotic-resistant C. jejuni as a serious antibiotic resistance threat in the United States. We recently identified a C. jejuni resistance enhancing CmeB (RE-CmeB) variant that can increase its multidrug efflux pump activity and confers an exceedingly high-level of resistance to fluoroquinolones. Here, we report the cryo-EM structures of this prevalent and clinically important C. jejuni RE-CmeB multidrug efflux pump in both the absence and presence of four antibiotics. These structures allow us to understand the action mechanism for multidrug recognition in this pump. Our studies will ultimately inform an era in structure-guided drug design to combat multidrug resistance in these Gram-negative pathogens. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40179.map.gz emd_40179.map.gz | 83.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40179-v30.xml emd-40179-v30.xml emd-40179.xml emd-40179.xml | 14.3 KB 14.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40179_fsc.xml emd_40179_fsc.xml | 13 KB | Display |  FSC data file FSC data file |
| Images |  emd_40179.png emd_40179.png | 162.1 KB | ||
| Filedesc metadata |  emd-40179.cif.gz emd-40179.cif.gz | 5.9 KB | ||
| Others |  emd_40179_half_map_1.map.gz emd_40179_half_map_1.map.gz emd_40179_half_map_2.map.gz emd_40179_half_map_2.map.gz | 154.5 MB 154.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40179 http://ftp.pdbj.org/pub/emdb/structures/EMD-40179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40179 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40179 | HTTPS FTP |
-Validation report
| Summary document |  emd_40179_validation.pdf.gz emd_40179_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40179_full_validation.pdf.gz emd_40179_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_40179_validation.xml.gz emd_40179_validation.xml.gz | 20.5 KB | Display | |
| Data in CIF |  emd_40179_validation.cif.gz emd_40179_validation.cif.gz | 26.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40179 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40179 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40179 | HTTPS FTP |
-Related structure data
| Related structure data |  8gk4MC  8gjjC  8gjkC  8gjlC  8gk0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40179.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40179.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Multi-drug efflux pump RE-CmeB bound with Chloramphenicol | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half Map 1
| File | emd_40179_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half Map 2
| File | emd_40179_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half Map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RE-CmeB
| Entire | Name: RE-CmeB |
|---|---|
| Components |
|
-Supramolecule #1: RE-CmeB
| Supramolecule | Name: RE-CmeB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Efflux pump membrane transporter
| Macromolecule | Name: Efflux pump membrane transporter / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 114.122477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFSKFFIERP IFASVVAIII SIAGIIGLAN LPVEQYPSLT PPTVQVSATY TGADAQTIAS TVATPIEDAI NGVDNMIYMD STSSPGQMK LTVYFNIGTD PDQAAIDVNN RISAATAKLP EAVKKLGVTV RKSSSTILEV VSVYSEDSSM NDIDIYNYVS L NILDELKR ...String: MFSKFFIERP IFASVVAIII SIAGIIGLAN LPVEQYPSLT PPTVQVSATY TGADAQTIAS TVATPIEDAI NGVDNMIYMD STSSPGQMK LTVYFNIGTD PDQAAIDVNN RISAATAKLP EAVKKLGVTV RKSSSTILEV VSVYSEDSSM NDIDIYNYVS L NILDELKR IPGVGDASAI GNKNYSMRIW LEPDLLNKFG VTANDVINAV NDQNAQYATG KIGEEPVVNK SPQVISITMQ GR LQTPQEF ENIILRVNED KSFLRIKDVA KVEIGAEQYN STGRLNTSAA VPIIINLQSG ANAVNTAKLI NEKMQELSKN FPQ GLKYQI PYDTTIFVKA SIKEVIKTFV EALALVLVVM YLFLKNFKST IIPMIAVPVS LLGTFAVLYV LGFSINLLTL FALV LAIGI VVDDAIIVVE NIDRILHEDS NISVKDAAIK AMNEVSSPVI SIVLVLCAVF IPVSFISGFV GEIQRQFALT LAISV AISG FVALTLTPSL SALFLTRNES KPFYFIQKFN DFFDWSTSVF SSGVAYILKR TIRFVLVFCI MIGFIAYLFK IVPSSL VPS EDQGVIMSII NLPSGSSIHR TIEEVDTINK NATQMKEISS SVSLIGFDLF TSSLKENAAA VFFILKDWSQ REASSDQ II AQLFGQYAAD RNALSYFLNL PPIPGLSLTG GFEMYAQNKS GKDYDAIQQD VNKMLELART RKELANVRTT LDTSFPQY K LIIDRDKMKY YNLNMQDVFN TISATIGTYY VNDFPMLGKN FQVNIRALGD FRNTQDALKN IYIRSSDNKM IPLNSFLTL VRSAGPDDVK RFNLFPAALI QGDPAPGYTS GQAIDAIAEV AKQSLGDEYS IAWSGSAYQE VSSKGAGAYA FVLGMIFVFL ILAAQYERW LMPLAVITAV PFAVFGSILL VALRGFDNDI YFQTGLLLLI GLSAKNAILI IEFAMEERLK KGKSIFEAAI N AAKLRFRP IIMTSLAFTF GVLPMIFATG AGSASRHSLG TGLIGGMIAA STLAIFFVPL FFYLLENFNE WLDKKRGKVH E UniProtKB: CmeB |
-Macromolecule #2: CHLORAMPHENICOL
| Macromolecule | Name: CHLORAMPHENICOL / type: ligand / ID: 2 / Number of copies: 1 / Formula: CLM |
|---|---|
| Molecular weight | Theoretical: 323.129 Da |
| Chemical component information | 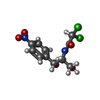 ChemComp-CLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 35.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)