+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of dimeric WDR11-FAM91A1 complex Body1 | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | Cryo-EM / Vesicle Trafficking / Neural Development / Protein Transport | |||||||||||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||||||||||||||
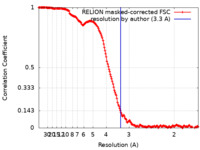
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||||||||
 Authors Authors | Jia GW / Deng QH / Su ZM / Jia D | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: The WDR11 complex is a receptor for acidic-cluster-containing cargo proteins. Authors: Huaqing Deng / Guowen Jia / Ping Li / Yingying Tang / Lin Zhao / Qin Yang / Jia Zhao / Jinrui Wang / Yingfeng Tu / Xin Yong / Sitao Zhang / Xianming Mo / Daniel D Billadeau / Zhaoming Su / Da Jia /   Abstract: Vesicle trafficking is a fundamental process that allows for the sorting and transport of specific proteins (i.e., "cargoes") to different compartments of eukaryotic cells. Cargo recognition ...Vesicle trafficking is a fundamental process that allows for the sorting and transport of specific proteins (i.e., "cargoes") to different compartments of eukaryotic cells. Cargo recognition primarily occurs through coats and the associated proteins at the donor membrane. However, it remains unclear whether cargoes can also be selected at other stages of vesicle trafficking to further enhance the fidelity of the process. The WDR11-FAM91A1 complex functions downstream of the clathrin-associated AP-1 complex to facilitate protein transport from endosomes to the TGN. Here, we report the cryo-EM structure of human WDR11-FAM91A1 complex. WDR11 directly and specifically recognizes a subset of acidic clusters, which we term super acidic clusters (SACs). WDR11 complex assembly and its binding to SAC-containing proteins are indispensable for the trafficking of SAC-containing proteins and proper neuronal development in zebrafish. Our studies thus uncover that cargo proteins could be recognized in a sequence-specific manner downstream of a protein coat. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39947.map.gz emd_39947.map.gz | 373.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39947-v30.xml emd-39947-v30.xml emd-39947.xml emd-39947.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39947_fsc.xml emd_39947_fsc.xml | 16.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_39947.png emd_39947.png | 74.5 KB | ||
| Filedesc metadata |  emd-39947.cif.gz emd-39947.cif.gz | 5.9 KB | ||
| Others |  emd_39947_half_map_1.map.gz emd_39947_half_map_1.map.gz emd_39947_half_map_2.map.gz emd_39947_half_map_2.map.gz | 306.9 MB 306.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39947 http://ftp.pdbj.org/pub/emdb/structures/EMD-39947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39947 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39947 | HTTPS FTP |
-Validation report
| Summary document |  emd_39947_validation.pdf.gz emd_39947_validation.pdf.gz | 731.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39947_full_validation.pdf.gz emd_39947_full_validation.pdf.gz | 730.9 KB | Display | |
| Data in XML |  emd_39947_validation.xml.gz emd_39947_validation.xml.gz | 24.7 KB | Display | |
| Data in CIF |  emd_39947_validation.cif.gz emd_39947_validation.cif.gz | 32.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39947 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39947 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39947 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39947 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39947.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39947.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size |
| ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39947_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of dimeric WDR11-FAM91A1 complex body1
| Entire | Name: Cryo-EM structure of dimeric WDR11-FAM91A1 complex body1 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of dimeric WDR11-FAM91A1 complex body1
| Supramolecule | Name: Cryo-EM structure of dimeric WDR11-FAM91A1 complex body1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Protein FAM91A1
| Macromolecule | Name: Protein FAM91A1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHHHHHHHHM NIDVEFHIRH NYPWNKLPAN VRQSLGNSQR EYEKQVVLYS IRNQLRYRNN LVKHVKKDER RYYEELLKYS RDHLMLYPY HLSDIMVKGL RITPFSYYTG IMEDIMNSEK SYDSLPNFTA ADCLRLLGIG RNQYIDLMNQ CRSSKKFFRR K TARDLLPI ...String: MHHHHHHHHM NIDVEFHIRH NYPWNKLPAN VRQSLGNSQR EYEKQVVLYS IRNQLRYRNN LVKHVKKDER RYYEELLKYS RDHLMLYPY HLSDIMVKGL RITPFSYYTG IMEDIMNSEK SYDSLPNFTA ADCLRLLGIG RNQYIDLMNQ CRSSKKFFRR K TARDLLPI KPVEIAIEAW WVVQAGYITE DDIKICTLPE KCAVDKIIDS GPQLSGSLDY NVVHSLYNKG FIYLDVPISD DS CIAVPPL EGFVMNRVQG DYFETLLYKI FVSIDEHTNV AELANVLEID LSLVKNAVSM YCRLGFAHKK GQVINLDQLH SSW KNVPSV NRLKSTLDPQ KMLLSWDGGE SRSPVQEASS ATDTDTNSQE DPADTASVSS LSLSTGHTKR IAFLFDSTLT AFLM MGNLS PNLKSHAVTM FEVGKLSDES LDSFLIELEK VQSTGEGEAQ RYFDHALTLR NTILFLRHNK DLVAQTAQPD QPNYG FPLD LLRCESLLGL DPATCSRVLN KNYTLLVSMA PLTNEIRPVS SCTPQHIGPA IPEVSSVWFK LYIYHVTGQG PPSLLL SKG TRLRKLPDIF QSYDRLLITS WGHDPGVVPT SNVLTMLNDA LTHSAVLIQG HGLHGIGETV HVPFPFDETE LQGEFTR VN MGVHKALQIL RNRVDLQHLC GYVTMLNASS QLADRKLSDA SDERGEPDLA SGSDVNGSTE SFEMVIEEAT IDSATKQT S GATTEADWVP LELCFGIPLF SSELNRKVCR KIAAHGLCRK ESLQNLLHSS RKLSLQVLNF VHSFQEGASI LDIHTEPSF SSLLSQSSCA DMGVPLPAKN LIFKDGVLSE WSGRSPSSLL IANLHLQ |
-Macromolecule #2: WD repeat-containing protein 11
| Macromolecule | Name: WD repeat-containing protein 11 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLPYTVNFKV SARTLTGALN AHNKAAVDWG WQGLIAYGCH SLVVVIDSIT AQTLQVLEKH KADVVKVKWA RENYHHNIGS PYCLRLASA DVNGKIIVWD VAAGVAQCEI QEHAKPIQDV QWLWNQDASR DLLLAIHPPN YIVLWNADTG TKLWKKSYAD N ILSFSFDP ...String: MLPYTVNFKV SARTLTGALN AHNKAAVDWG WQGLIAYGCH SLVVVIDSIT AQTLQVLEKH KADVVKVKWA RENYHHNIGS PYCLRLASA DVNGKIIVWD VAAGVAQCEI QEHAKPIQDV QWLWNQDASR DLLLAIHPPN YIVLWNADTG TKLWKKSYAD N ILSFSFDP FDPSHLTLLT SEGIVFISDF SPSKPPSGPG KKVYISSPHS SPAHNKLATA TGAKKALNKV KILITQEKPS AE FITLNDC LQLAYLPSKR NHMLLLYPRE ILILDLEVNQ TVGVIAIERT GVPFLQVIPC FQRDGLFCLH ENGCITLRVR RSY NNIFTT SNEEPDPDPV QELTYDLRSQ CDAIRVTKTV RPFSMVCCPV NENAAALVVS DGRVMIWELK SAVCNRNSRN SSSG VSPLY SPVSFCGIPV GVLQNKLPDL SLDNMIGQSA IAGEEHPRGS ILREVHLKFL LTGLLSGLPA PQFAIRMCPP LTTKN IKMY QPLLAVGTSN GSVLVYHLTS GLLHKELSIH SCEVKGIEWT SLTSFLSFAT STPNNMGLVR NELQLVDLPT GRSIAF RGE RGNDESAIEM IKVSHLKQYL AVVFRDKPLE LWDVRTCTLL REMSKNFPTI TALEWSPSHN LKSLRKKQLA TREAMAR QT VVSDTELSIV ESSVISLLQE AESKSELSQN ISAREHFVFT DIDGQVYHLT VEGNSVKDSA RIPPDGSMGS ITCIAWKG D TLVLGDMDGN LNFWDLKGRV SRGIPTHRSW VRKIRFAPGK GNQKLIAMYN DGAEVWDTKE VQMVSSLRSG RNVTFRILD VDWCTSDKVI LASDDGCIRV LEMSMKSACF RMDEQELTEP VWCPYLLVPR ASLALKAFLL HQPWNGQYSL DISHVDYPEN EEIKNLLQE QLNSLSNDIK KLLLDPEFTL LQRCLLVSRL YGDESELHFW TVAAHYLHSL SQEKSASTTA PKEAAPRDKL S NPLDICYD VLCENAYFQK FQLERVNLQE VKRSTYDHTR KCTDQLLLLG QTDRAVQLLL ETSADNQHYY CDSLKACLVT TV TSSGPSQ STIKLVATNM IANGKLAEGV QLLCLIDKAA DACRYLQTYG EWNRAAWLAK VRLNPEECAD VLRRWVDHLC SPQ VNQKSK ALLVLLSLGC FFSVAETLHS MRYFDRAALF VEACLKYGAF EVTEDTEKLI TAIYADYARS LKNLGFKQGA VLFA SKAGA AGKDLLNELE SPKEEPIEE |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 58.47 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.992 µm / Nominal defocus min: 0.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)