+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of HBMBPP-BTN2A1-BTN3A1 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / immune / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationButyrophilin (BTN) family interactions / activated T cell proliferation / regulation of cytokine production / positive regulation of cytokine production / lipid metabolic process / positive regulation of type II interferon production / T cell receptor signaling pathway / adaptive immune response / external side of plasma membrane / signaling receptor binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Zheng J / Gao W / Zhu Y / Huang Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of HBMBPP-BTN2A1-BTN3A1 complex Authors: Zheng J / Gao W / Zhu Y / Huang Z | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39875.map.gz emd_39875.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39875-v30.xml emd-39875-v30.xml emd-39875.xml emd-39875.xml | 15.7 KB 15.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39875.png emd_39875.png | 86.4 KB | ||
| Filedesc metadata |  emd-39875.cif.gz emd-39875.cif.gz | 5.9 KB | ||
| Others |  emd_39875_half_map_1.map.gz emd_39875_half_map_1.map.gz emd_39875_half_map_2.map.gz emd_39875_half_map_2.map.gz | 226.4 MB 226.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39875 http://ftp.pdbj.org/pub/emdb/structures/EMD-39875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39875 | HTTPS FTP |
-Validation report
| Summary document |  emd_39875_validation.pdf.gz emd_39875_validation.pdf.gz | 751.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39875_full_validation.pdf.gz emd_39875_full_validation.pdf.gz | 751.4 KB | Display | |
| Data in XML |  emd_39875_validation.xml.gz emd_39875_validation.xml.gz | 15.5 KB | Display | |
| Data in CIF |  emd_39875_validation.cif.gz emd_39875_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39875 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39875 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39875 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39875 | HTTPS FTP |
-Related structure data
| Related structure data |  8za9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39875.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39875.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0773 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39875_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_39875_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BTN2A1-BTN3A1
| Entire | Name: BTN2A1-BTN3A1 |
|---|---|
| Components |
|
-Supramolecule #1: BTN2A1-BTN3A1
| Supramolecule | Name: BTN2A1-BTN3A1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Butyrophilin subfamily 3 member A1
| Macromolecule | Name: Butyrophilin subfamily 3 member A1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 54.432809 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QFSVLGPSGP ILAMVGEDAD LPCHLFPTMS AETMELKWVS SSLRQVVNVY ADGKEVEDRQ SAPYRGRTSI LRDGITAGKA ALRIHNVTA SDSGKYLCYF QDGDFYEKAL VELKVAALGS DLHVDVKGYK DGGIHLECRS TGWYPQPQIQ WSNNKGENIP T VEAPVVAD ...String: QFSVLGPSGP ILAMVGEDAD LPCHLFPTMS AETMELKWVS SSLRQVVNVY ADGKEVEDRQ SAPYRGRTSI LRDGITAGKA ALRIHNVTA SDSGKYLCYF QDGDFYEKAL VELKVAALGS DLHVDVKGYK DGGIHLECRS TGWYPQPQIQ WSNNKGENIP T VEAPVVAD GVGLYAVAAS VIMRGSSGEG VSCTIRSSLL GLEKTASISI ADPFFRSAQR WIAALAGTLP VLLLLLGGAG YF LWQQQEE KKTQFRKKKR EQELREMAWS TMKQEQSTRV KLLEELRWRS IQYASRGERH SAYNEWKKAL FKPADVILDP KTA NPILLV SEDQRSVQRA KEPQDLPDNP ERFNWHYCVL GCESFISGRH YWEVEVGDRK EWHIGVCSKN VQRKGWVKMT PENG FWTMG LTDGNKYRTL TEPRTNLKLP KTPKKVGVFL DYETGDISFY NAVDGSHIHT FLDVSFSEAL YPVFRILTLE PTALT ICPA UniProtKB: Butyrophilin subfamily 3 member A1 |
-Macromolecule #2: Butyrophilin subfamily 2 member A1
| Macromolecule | Name: Butyrophilin subfamily 2 member A1 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.826953 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QFIVVGPTDP ILATVGENTT LRCHLSPEKN AEDMEVRWFR SQFSPAVFVY KGGRERTEEQ MEEYRGRTTF VSKDISRGSV ALVIHNITA QENGTYRCYF QEGRSYDEAI LHLVVAGLGS KPLISMRGHE DGGIRLECIS RGWYPKPLTV WRDPYGGVAP A LKEVSMPD ...String: QFIVVGPTDP ILATVGENTT LRCHLSPEKN AEDMEVRWFR SQFSPAVFVY KGGRERTEEQ MEEYRGRTTF VSKDISRGSV ALVIHNITA QENGTYRCYF QEGRSYDEAI LHLVVAGLGS KPLISMRGHE DGGIRLECIS RGWYPKPLTV WRDPYGGVAP A LKEVSMPD ADGLFMVTTA VIIRDKSVRN MSCSINNTLL GQKKESVIFI PESFMPSVSP CAVALPIIVV ILMIPIAVCI YW INKLQKE KKILSGEKEF ERETREIALK ELEKERVQKE EELQVKEKLQ EELRWRRTFL HAVDVVLDPD TAHPDLFLSE DRR SVRRCP FRHLGESVPD NPERFDSQPC VLGRESFASG KHYWEVEVEN VIEWTVGVCR DSVERKGEVL LIPQNGFWTL EMHK GQYRA VSSPDRILPL KESLCRVGVF LDYEAGDVSF YNMRDRSHIY TCPRSAFSVP VRPFFRLGCE DSPIFICPAL TGANG VTVP EEGLTLHRVG THQSL UniProtKB: Butyrophilin subfamily 2 member A1 |
-Macromolecule #3: (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate
| Macromolecule | Name: (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate type: ligand / ID: 3 / Number of copies: 1 / Formula: H6P |
|---|---|
| Molecular weight | Theoretical: 262.092 Da |
| Chemical component information | 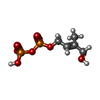 ChemComp-H6P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: NITROGEN |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 50 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 535747 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)