[English] 日本語
 Yorodumi
Yorodumi- EMDB-39850: Cryo-EM structure of Thogoto virus polymerase in a transcription ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationcap snatching / 7-methylguanosine mRNA capping / virion component / RNA-directed RNA polymerase / viral RNA genome replication / RNA-dependent RNA polymerase activity / nucleotide binding / host cell nucleus / RNA binding Similarity search - Function | |||||||||
| Biological species |  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) | |||||||||
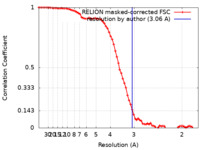
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Xue L / Chang T / Li Z / Zhao H / Li M / He J / Chen X / Xiong X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses. Authors: Lu Xue / Tiancai Chang / Zimu Li / Chenchen Wang / Heyu Zhao / Mei Li / Peng Tang / Xin Wen / Mengmeng Yu / Jiqin Wu / Xichen Bao / Xiaojun Wang / Peng Gong / Jun He / Xinwen Chen / Xiaoli Xiong /  Abstract: Influenza viruses and thogotoviruses account for most recognized orthomyxoviruses. Thogotoviruses, exemplified by Thogoto virus (THOV), are capable of infecting humans using ticks as vectors. THOV ...Influenza viruses and thogotoviruses account for most recognized orthomyxoviruses. Thogotoviruses, exemplified by Thogoto virus (THOV), are capable of infecting humans using ticks as vectors. THOV transcribes mRNA without the extraneous 5' end sequences derived from cap-snatching in influenza virus mRNA. Here, we report cryo-EM structures to characterize THOV polymerase RNA synthesis initiation and elongation. The structures demonstrate that THOV RNA transcription and replication are able to start with short dinucleotide primers and that the polymerase cap-snatching machinery is likely non-functional. Triggered by RNA synthesis, asymmetric THOV polymerase dimers can form without the involvement of host factors. We confirm that, distinctive from influenza viruses, THOV-polymerase RNA synthesis is weakly dependent of the host factors ANP32A/B/E in human cells. This study demonstrates varied mechanisms in RNA synthesis and host factor utilization among orthomyxoviruses, providing insights into the mechanisms behind thogotoviruses' broad-infectivity range. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39850.map.gz emd_39850.map.gz | 23.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39850-v30.xml emd-39850-v30.xml emd-39850.xml emd-39850.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_39850_fsc.xml emd_39850_fsc.xml | 7.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_39850.png emd_39850.png | 24.2 KB | ||
| Masks |  emd_39850_msk_1.map emd_39850_msk_1.map | 34.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-39850.cif.gz emd-39850.cif.gz | 7.2 KB | ||
| Others |  emd_39850_half_map_1.map.gz emd_39850_half_map_1.map.gz emd_39850_half_map_2.map.gz emd_39850_half_map_2.map.gz | 26.4 MB 26.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39850 http://ftp.pdbj.org/pub/emdb/structures/EMD-39850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39850 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39850 | HTTPS FTP |
-Validation report
| Summary document |  emd_39850_validation.pdf.gz emd_39850_validation.pdf.gz | 981.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39850_full_validation.pdf.gz emd_39850_full_validation.pdf.gz | 980.8 KB | Display | |
| Data in XML |  emd_39850_validation.xml.gz emd_39850_validation.xml.gz | 13.5 KB | Display | |
| Data in CIF |  emd_39850_validation.cif.gz emd_39850_validation.cif.gz | 18.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39850 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39850 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39850 | HTTPS FTP |
-Related structure data
| Related structure data |  8z8xMC  8z85C  8z8jC  8z8nC  8z90C  8z97C  8z98C  8z9hC  8z9qC  8z9rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39850.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39850.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.89846 Å | ||||||||||||||||||||||||||||||||||||
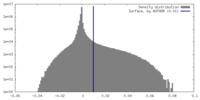
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_39850_msk_1.map emd_39850_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
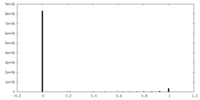
| Density Histograms |
-Half map: #2
| File | emd_39850_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
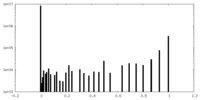
| Density Histograms |
-Half map: #1
| File | emd_39850_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
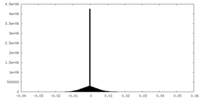
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of Thogoto virus polymerase in a transcription ...
| Entire | Name: Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of Thogoto virus polymerase in a transcription ...
| Supramolecule | Name: Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
-Macromolecule #1: Polymerase acidic protein
| Macromolecule | Name: Polymerase acidic protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
| Molecular weight | Theoretical: 71.623445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDRPDHIDS RVWELSETQE DWITQVHGHV RRVVECWKYT ICCLISNMHT HRGAPQYDVF KWQDRSTIEW ICSKKKVQYP ERDTPDLYD NERAVAYKVL LVSDLSDHSP TSGIYHDLAF NLEGEAEESC ALVLRGSQLQ DIKGFLCRAL EWVVSNNLTQ E VVETISGE ...String: MTDRPDHIDS RVWELSETQE DWITQVHGHV RRVVECWKYT ICCLISNMHT HRGAPQYDVF KWQDRSTIEW ICSKKKVQYP ERDTPDLYD NERAVAYKVL LVSDLSDHSP TSGIYHDLAF NLEGEAEESC ALVLRGSQLQ DIKGFLCRAL EWVVSNNLTQ E VVETISGE AKLQFSVGTT FRTLLKRDTD WDVIPTPRVE PNVPRIEGRR WTQMKKLPLL KEKEGPPSPW RALLLGADSE YI VCPPGTD QEAISWIHSQ SEIECIRESK STPASVITCL TSSLQSFAEG NPVRSRIHED IIAFGINKKQ EKKQSASSSA SGE WKRAEY QVEEMSLPPW VEEEMVLLRS DQEDNWIELE KNAIYTEVDG VAEGLVDKYI EIVGRTKVAS VIEKWQIAAT RTFS QLHTD RSRITACPII TRDPSGNCQF WGMVLLGPHH VKRDTDNAPL LIAEIMGEDT EEKYPKHSVF SLKVEEKQFL LSLKI TSFS RNKLYTFSNI RRVLIQPASI YSQVVLSRAA ENNSLNLEVN PEIQLYLEGA QRGMTLYQWV RMILCLEFLM AIYNNP QME GFLANMRRLH MSRHAMMERR QVFLPFGSRP EDKVNECIIN NPIVAYLAKG WNSMPNVYY UniProtKB: Polymerase acidic protein |
-Macromolecule #2: RNA-directed RNA polymerase catalytic subunit
| Macromolecule | Name: RNA-directed RNA polymerase catalytic subunit / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: RNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
| Molecular weight | Theoretical: 81.432664 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNLFTPLSEI NPTTTQELLY AYTGPAPVAY GTRTRAVLEN IIRPYQYFYK EPNVQRALDI KTGCKEPEDI NVEGPSSGFH TASVLKLAD NFFRKYRPAM EKLKYWILVK LPKLKYAELS KGRQTYSFIH KRNLPAPIAL EETVEFLEQN LRRKIGPTLL S YCQAIADV ...String: MNLFTPLSEI NPTTTQELLY AYTGPAPVAY GTRTRAVLEN IIRPYQYFYK EPNVQRALDI KTGCKEPEDI NVEGPSSGFH TASVLKLAD NFFRKYRPAM EKLKYWILVK LPKLKYAELS KGRQTYSFIH KRNLPAPIAL EETVEFLEQN LRRKIGPTLL S YCQAIADV MELDETTYEG ARDPRPWDIQ LEEIDSDEED PLFRQVGREE TYTIKFSREE LWDQMRTLNT MWKHLERGRL NR RTIATPS MLIRGFVKIV EDAAKEILEN VPTSGVPVGG EEKLAKLASK QTFHTAVTGE LSGDQEKFNE CLDPDAMRLM WTV FLRKLG CPDWIMELFN IPFMVFKSKL ADMGEGLVYT KGKLTDRKPL GEMPSEFDDL VRNVVGNSIS CRLGMFMGMY NLTS TLLAL ISIEREELTG SHVESSDDFI HFFNCKTHEE MFKQAETLRL TLKLVGINMS PSKCILISPA GIGEFNSKFH HRDFV GNVA TELPALVPNG TNPMTDLAMG LNVIKHSVNT GQMNLCTGAL AMRIFNHAYK YAYMALGVTR RTRFMEENAI TPLLTN QGA SPVHSFSTMH LDEVALRRHL GLLDEETLRR ILNPNNPVTQ KGDPSMFFRI ENKMPQIMED YSVPSCFKYT LSRNRTI QD KPHKALLNKE ERYQRVTSII NKLFPEVLIQ EASAPGTVRE SLKRRLELVV ERSDLDEERK KRILSRIF UniProtKB: RNA-directed RNA polymerase catalytic subunit |
-Macromolecule #3: Polymerase basic protein 2
| Macromolecule | Name: Polymerase basic protein 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
| Molecular weight | Theoretical: 94.418578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDREEPAESE CTLRALVEEY NGACKEAPKE MSKQFTDYNT FKRYTTSKKD HAPQMRLVYS VRKPWPISMT PSKEIPLVFN GTKLKDTIL DLGESKRTRA NIVVPDYWSK YGSQTSLEVV NAILYAEDLK VQRFFSTEWG EIRYGRMLPF RKPVQACPTI E EVNPASIP ...String: MDREEPAESE CTLRALVEEY NGACKEAPKE MSKQFTDYNT FKRYTTSKKD HAPQMRLVYS VRKPWPISMT PSKEIPLVFN GTKLKDTIL DLGESKRTRA NIVVPDYWSK YGSQTSLEVV NAILYAEDLK VQRFFSTEWG EIRYGRMLPF RKPVQACPTI E EVNPASIP HTLLQVFCPQ YTTLDSKRKA HMGAVEKLKR VMEPICKVQT QESAVHIARS LIDSNKKWLP TVVDHTPRTA EM AHFLCSK YHYVHTNTQD LSDTRSIDNL CGELVKRSLK CRCPKETLVA NLDKITIQGR PMREVLADHD GELPYLGICR VAM GLSTHH TMKIRSTKFS ILNSDHPRIE VKKVFSLSPD VQVTIPYRRF KGKAKVYFQN DQIQGYFSCT DRQIDEIKIS APKN APLLE PLLDICYYGS FIEPGFEQTF GFYPAGKREF VDSFFMHHSK DHKAFLIHMG LDKDLSLPLS PELNWKEPAL SKVCR VTEL DSTVQPYTSA TREFVLGETL NVYTQHENGL ELLICPTEIR STRGPLPPGT NLSGSEFIDI YQDPFSRAKS LLKSTI LHA ERCKEFVGNM LEEYQDPAET TVQSLVPINT WGKSAKRKLQ EEITSDPDWH QCPRKRAKMS YLAIIAGSIQ DRDKKQT NV PRAFMLRGSQ IEYDMKATRG LVVDTTNRII VGGETVLREG KGGPEGYVQT GVFEEQPRCY LVDTPDHGLS MGLSRFCV H SQGRYFQYEK KISIWEETDN IKATIDSQRD LKRRRDIEEM VSKRARIVLE VLFQGPGHHH HHHHHSADYK DDDDKGGWS HPQFEKGGGS GGGGSGGSAW SHPQFEK UniProtKB: Polymerase basic protein 2 |
-Macromolecule #4: RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3') type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
| Molecular weight | Theoretical: 5.84358 KDa |
| Sequence | String: AGAGAAAUCA AGGCAGUU |
-Macromolecule #5: RNA (5'-R(*GP*AP*CP*UP*GP*CP*CP*UP*GP*UP*UP*UP*UP*UP*GP*CP*U)-3')
| Macromolecule | Name: RNA (5'-R(*GP*AP*CP*UP*GP*CP*CP*UP*GP*UP*UP*UP*UP*UP*GP*CP*U)-3') type: rna / ID: 5 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Thogoto virus (isolate SiAr 126) Thogoto virus (isolate SiAr 126) |
| Molecular weight | Theoretical: 5.335125 KDa |
| Sequence | String: GACUGCCUGU UUUUGCU |
-Macromolecule #6: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER
| Macromolecule | Name: PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: G2P |
|---|---|
| Molecular weight | Theoretical: 521.208 Da |
| Chemical component information |  ChemComp-G2P: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: TFS FALCON 4i (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)