+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the immepip-bound H3R-Gi complex | |||||||||
 Map data Map data | Sharpened Map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / H3R / Histamine Receptor / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationHistamine receptors / histamine receptor activity / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / neurotransmitter secretion / G protein-coupled serotonin receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / T cell migration / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway ...Histamine receptors / histamine receptor activity / G protein-coupled acetylcholine receptor activity / adenylate cyclase-inhibiting G protein-coupled acetylcholine receptor signaling pathway / neurotransmitter secretion / G protein-coupled serotonin receptor activity / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / T cell migration / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / G protein-coupled serotonin receptor binding / cellular response to forskolin / regulation of mitotic spindle organization / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Regulation of insulin secretion / G protein-coupled receptor binding / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / response to peptide hormone / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / cellular response to catecholamine stimulus / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / presynapse / retina development in camera-type eye / Ca2+ pathway / cell cortex / phospholipase C-activating G protein-coupled receptor signaling pathway / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / chemical synaptic transmission / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / cell division / GTPase activity / centrosome / dendrite / synapse / protein-containing complex binding / GTP binding / nucleolus / magnesium ion binding / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Shen Q / Tang X / Wen X / Cheng S / Xiao P / Zang S / Shen D / Jiang L / Zheng Y / Zhang H ...Shen Q / Tang X / Wen X / Cheng S / Xiao P / Zang S / Shen D / Jiang L / Zheng Y / Zhang H / Xu H / Mao C / Zhang M / Hu W / Sun J / Chen Z / Zhang Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Adv Sci (Weinh) / Year: 2024 Journal: Adv Sci (Weinh) / Year: 2024Title: Molecular Determinant Underlying Selective Coupling of Primary G-Protein by Class A GPCRs. Authors: Qingya Shen / Xinyan Tang / Xin Wen / Shizhuo Cheng / Peng Xiao / Shao-Kun Zang / Dan-Dan Shen / Lei Jiang / Yanrong Zheng / Huibing Zhang / Haomang Xu / Chunyou Mao / Min Zhang / Weiwei Hu ...Authors: Qingya Shen / Xinyan Tang / Xin Wen / Shizhuo Cheng / Peng Xiao / Shao-Kun Zang / Dan-Dan Shen / Lei Jiang / Yanrong Zheng / Huibing Zhang / Haomang Xu / Chunyou Mao / Min Zhang / Weiwei Hu / Jin-Peng Sun / Yan Zhang / Zhong Chen /  Abstract: G-protein-coupled receptors (GPCRs) transmit downstream signals predominantly via G-protein pathways. However, the conformational basis of selective coupling of primary G-protein remains elusive. ...G-protein-coupled receptors (GPCRs) transmit downstream signals predominantly via G-protein pathways. However, the conformational basis of selective coupling of primary G-protein remains elusive. Histamine receptors HR and HR couple with G- or G-proteins respectively. Here, three cryo-EM structures of HR-G and HR-G complexes are presented at a global resolution of 2.6-2.7 Å. These structures reveal the unique binding pose for endogenous histamine in HR, wherein the amino group interacts with E206 of HR instead of the conserved D114 of other aminergic receptors. Furthermore, comparative analysis of the HR-G and HR-G complexes reveals that the structural geometry of TM5/TM6 determines the primary G-protein selectivity in histamine receptors. Machine learning (ML)-based structuromic profiling and functional analysis of class A GPCR-G-protein complexes illustrate that TM5 length, TM5 tilt, and TM6 outward movement are key determinants of the G and G selectivity among the whole Class A family. Collectively, the findings uncover the common structural geometry within class A GPCRs that determines the primary G- and G-coupling selectivity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39584.map.gz emd_39584.map.gz | 17.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39584-v30.xml emd-39584-v30.xml emd-39584.xml emd-39584.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39584.png emd_39584.png | 154.9 KB | ||
| Filedesc metadata |  emd-39584.cif.gz emd-39584.cif.gz | 6.7 KB | ||
| Others |  emd_39584_half_map_1.map.gz emd_39584_half_map_1.map.gz emd_39584_half_map_2.map.gz emd_39584_half_map_2.map.gz | 20.7 MB 20.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39584 http://ftp.pdbj.org/pub/emdb/structures/EMD-39584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39584 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39584 | HTTPS FTP |
-Validation report
| Summary document |  emd_39584_validation.pdf.gz emd_39584_validation.pdf.gz | 768.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39584_full_validation.pdf.gz emd_39584_full_validation.pdf.gz | 767.9 KB | Display | |
| Data in XML |  emd_39584_validation.xml.gz emd_39584_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_39584_validation.cif.gz emd_39584_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39584 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39584 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39584 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39584 | HTTPS FTP |
-Related structure data
| Related structure data |  8yuvMC  8yutC  8yuuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39584.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39584.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened Map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 2
| File | emd_39584_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
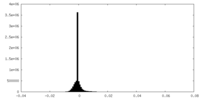
| Density Histograms |
-Half map: Half map 1
| File | emd_39584_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
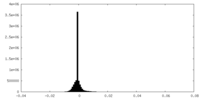
| Density Histograms |
- Sample components
Sample components
-Entire : Histamine-bound H3R-Gi complex
| Entire | Name: Histamine-bound H3R-Gi complex |
|---|---|
| Components |
|
-Supramolecule #1: Histamine-bound H3R-Gi complex
| Supramolecule | Name: Histamine-bound H3R-Gi complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.414047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKNTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVGAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.418086 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR ...String: MHHHHHHLEV LFQGPGSSGS ELDQLRQEAE QLKNQIRDAR KACADATLSQ ITNNIDPVGR IQMRTRRTLR GHLAKIYAMH WGTDSRLLV SASQDGKLII WDSYTTNKVH AIPLRSSWVM TCAYAPSGNY VACGGLDNIC SIYNLKTREG NVRVSRELAG H TGYLSCCR FLDDNQIVTS SGDTTCALWD IETGQQTTTF TGHTGDVMSL SLAPDTRLFV SGACDASAKL WDVREGMCRQ TF TGHESDI NAICFFPNGN AFATGSDDAT CRLFDLRADQ ELMTYSHDNI ICGITSVSFS KSGRLLLAGY DDFNCNVWDA LKA DRAGVL AGHDNRVSCL GVTDDGMAVA TGSWDSFLKI WN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 28.813047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL KGSLEVLFQG PAAAHHHHHH HH |
-Macromolecule #5: Histamine H3 receptor
| Macromolecule | Name: Histamine H3 receptor / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.603223 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DYKDDDDKLE VLFQGPMERA PPDGPLNASG ALAGEAAAAG GARGFSAAWT AVLAALMALL IVATVLGNAL VMLAFVADSS LRTQNNFFL LNLAISDFLV GAFCIPLYVP YVLTGRWTFG RGLCKLWLVV DYLLCTSSAF NIVLISYDRF LSVTRAVSYR A QQGDTRRA ...String: DYKDDDDKLE VLFQGPMERA PPDGPLNASG ALAGEAAAAG GARGFSAAWT AVLAALMALL IVATVLGNAL VMLAFVADSS LRTQNNFFL LNLAISDFLV GAFCIPLYVP YVLTGRWTFG RGLCKLWLVV DYLLCTSSAF NIVLISYDRF LSVTRAVSYR A QQGDTRRA VRKMLLVWVL AFLLYGPAIL SWEYLSGGSS IPEGHCYAEF FYNWYFLITA STLEFFTPFL SVTFFNLSIY LN IQRRTRL RLDGAREAAG PEPPPEAQPS PPPPPGCWGC WQKGHGEAMP LHRYGVGEAA VGAEAGEATL GGGGGGGSVA SPT SSSGSS SRGTERPRSL KRGSKPSASS ASLEKRMKMV SQSFTQRFRL SRDRKVAKSL AVIVSIFGLC WAPYTLLMII RAAC HGHCV PDYWYETSFW LLWANSAVNP VLYPLCHHSF RRAFTKLLCP QKLKIQPHSS LEHCWK UniProtKB: Histamine H3 receptor |
-Macromolecule #6: 4-(1H-imidazol-5-ylmethyl)piperidine
| Macromolecule | Name: 4-(1H-imidazol-5-ylmethyl)piperidine / type: ligand / ID: 6 / Number of copies: 1 / Formula: A1LY4 |
|---|---|
| Molecular weight | Theoretical: 165.235 Da |
-Macromolecule #7: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 7 / Number of copies: 1 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 62.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 330449 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-8yuv: |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)