+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | OSCA1.1-F516A open | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | OSCA1.1-F516A open / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of calcium ion import / calcium-activated cation channel activity / cellular hyperosmotic response / response to osmotic stress / monoatomic cation channel activity / protein tetramerization / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zhang MF | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Activation mechanisms of dimeric mechanosensitive OSCA/TMEM63 channels. Authors: Yuanyue Shan / Mengmeng Zhang / Meiyu Chen / Xinyi Guo / Ying Li / Mingfeng Zhang / Duanqing Pei /  Abstract: OSCA/TMEM63 channels, which have transporter-like architectures, are bona fide mechanosensitive (MS) ion channels that sense high-threshold mechanical forces in eukaryotic cells. The activation ...OSCA/TMEM63 channels, which have transporter-like architectures, are bona fide mechanosensitive (MS) ion channels that sense high-threshold mechanical forces in eukaryotic cells. The activation mechanism of these transporter-like channels is not fully understood. Here we report cryo-EM structures of a dimeric OSCA/TMEM63 pore mutant OSCA1.1-F516A with a sequentially extracellular dilated pore in a detergent environment. These structures suggest that the extracellular pore sequential dilation resembles a flower blooming and couples to a sequential contraction of each monomer subunit towards the dimer interface and subsequent extrusion of the dimer interface lipids. Interestingly, while OSCA1.1-F516A remains non-conducting in the native lipid environment, it can be directly activated by lyso-phosphatidylcholine (Lyso-PC) with reduced single-channel conductance. Structural analysis of OSCA1.1-F516A in lyso-PC-free and lyso-PC-containing lipid nanodiscs indicates that lyso-PC induces intracellular pore dilation by attracting the M6b to upward movement away from the intracellular side thus extending the intracellular pore. Further functional studies indicate that full activation of MS OSCA/TMEM63 dimeric channels by high-threshold mechanical force also involves the opening of both intercellular and extracellular pores. Our results provide the fundamental activation paradigm of the unique transporter-like MS OSCA/TMEM63 channels, which is likely applicable to functional branches of the TMEM63/TMEM16/TMC superfamilies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39399.map.gz emd_39399.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39399-v30.xml emd-39399-v30.xml emd-39399.xml emd-39399.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39399.png emd_39399.png | 54.7 KB | ||
| Filedesc metadata |  emd-39399.cif.gz emd-39399.cif.gz | 5.6 KB | ||
| Others |  emd_39399_half_map_1.map.gz emd_39399_half_map_1.map.gz emd_39399_half_map_2.map.gz emd_39399_half_map_2.map.gz | 1.3 GB 1.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39399 http://ftp.pdbj.org/pub/emdb/structures/EMD-39399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39399 | HTTPS FTP |
-Validation report
| Summary document |  emd_39399_validation.pdf.gz emd_39399_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39399_full_validation.pdf.gz emd_39399_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_39399_validation.xml.gz emd_39399_validation.xml.gz | 23.5 KB | Display | |
| Data in CIF |  emd_39399_validation.cif.gz emd_39399_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39399 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39399 | HTTPS FTP |
-Related structure data
| Related structure data |  8ymmMC  8ymnC  8ymoC  8ympC  8ymqC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_39399.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39399.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.35 Å | ||||||||||||||||||||||||||||||||||||
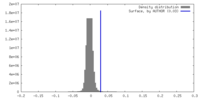
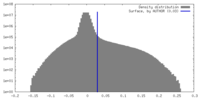
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39399_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
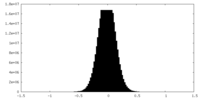
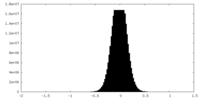
| Density Histograms |
-Half map: #1
| File | emd_39399_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
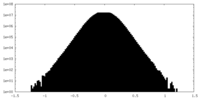
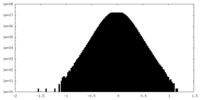
| Density Histograms |
- Sample components
Sample components
-Entire : human PIEZO1
| Entire | Name: human PIEZO1 |
|---|---|
| Components |
|
-Supramolecule #1: human PIEZO1
| Supramolecule | Name: human PIEZO1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Protein OSCA1
| Macromolecule | Name: Protein OSCA1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 87.620914 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATLKDIGVS AGINILTAFI FFIIFAFLRL QPFNDRVYFS KWYLRGLRSS PASGGGFAGR FVNLELRSYL KFLHWMPEAL KMPERELID HAGLDSVVYL RIYWLGLKIF APIAMLAWAV LVPVNWTNNE LELAKHFKNV TSSDIDKLTI SNIPEGSNRF W AHIIMAYA ...String: MATLKDIGVS AGINILTAFI FFIIFAFLRL QPFNDRVYFS KWYLRGLRSS PASGGGFAGR FVNLELRSYL KFLHWMPEAL KMPERELID HAGLDSVVYL RIYWLGLKIF APIAMLAWAV LVPVNWTNNE LELAKHFKNV TSSDIDKLTI SNIPEGSNRF W AHIIMAYA FTIWTCYMLM KEYETVANMR LQFLASEGRR PDQFTVLVRN VPPDPDETVS ELVEHFFLVN HPDNYLTHQV VC NANKLAD LVSKKTKLQN WLDYYQLKYT RNNSQIRPIT KLGCLGLCGQ KVDAIEHYIA EVDKTSKEIA EERENVVNDQ KSV MPASFV SFKTRWAAAV CAQTTQTRNP TEWLTEWAAE PRDIYWPNLA IPYVSLTVRR LVMNVAFFFL TFFFIIPIAF VQSL ATIEG IEKVAPFLKV IIEKDFIKSL IQGLLAGIAL KLFLIFLPAI LMTMSKFEGF TSVSFLERRS ASRYYIFNLV NVFLG SVIA GAAFEQLNSF LNQSPNQIPK TIGMAIPMKA TAFITYIMVD GWAGVAGEIL MLKPLIIYHL KNAFLVKTEK DREEAM NPG SIGFNTGEPQ IQLYFLLGLV YAPVTPMLLP FILVFFALAY VVYRHQIINV YNQEYESAAA FWPDVHGRVI TALIISQ LL LMGLLGTKHA ASAAPFLIAL PVITIGFHRF CKGRFEPAFV RYPLQEAMMK DTLERAREPN LNLKGYLQDA YIHPVFKG G DNDDDGDMIG KLENEVIIVP TKRQSRRNTP APSRISGESS PSLAVINGKE V UniProtKB: Protein OSCA1 |
-Macromolecule #2: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
| Macromolecule | Name: 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine / type: ligand / ID: 2 / Number of copies: 2 / Formula: PEE |
|---|---|
| Molecular weight | Theoretical: 744.034 Da |
| Chemical component information |  ChemComp-PEE: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 54900 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)