[English] 日本語
 Yorodumi
Yorodumi- EMDB-39036: Structure of HCoV-HKU1C spike in the functionally anchored-1up co... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HKU1A / spike / TMPRSS2 / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransmembrane protease serine 2 / symbiont-mediated induction of syncytium formation / protein autoprocessing / Attachment and Entry / serine-type peptidase activity / viral translation / Induction of Cell-Cell Fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / Attachment and Entry / positive regulation of viral entry into host cell ...transmembrane protease serine 2 / symbiont-mediated induction of syncytium formation / protein autoprocessing / Attachment and Entry / serine-type peptidase activity / viral translation / Induction of Cell-Cell Fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / host cell plasma membrane / virion membrane / proteolysis / extracellular exosome / extracellular region / nucleoplasm / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Human coronavirus HKU1 / Human coronavirus HKU1 /  Human coronavirus HKU1 (isolate N5) / Human coronavirus HKU1 (isolate N5) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.26 Å | |||||||||
 Authors Authors | Lu YC / Zhang X / Wang HF / Liu XC / Sun L / Yang HT | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2024 Journal: Cell / Year: 2024Title: TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry. Authors: Haofeng Wang / Xiaoce Liu / Xiang Zhang / Zhuoqian Zhao / Yuchi Lu / Dingzhe Pu / Zeyang Zhang / Jie Chen / Yajie Wang / Mengfei Li / Xuxue Dong / Yinkai Duan / Yujia He / Qiyu Mao / ...Authors: Haofeng Wang / Xiaoce Liu / Xiang Zhang / Zhuoqian Zhao / Yuchi Lu / Dingzhe Pu / Zeyang Zhang / Jie Chen / Yajie Wang / Mengfei Li / Xuxue Dong / Yinkai Duan / Yujia He / Qiyu Mao / Hangtian Guo / Haoran Sun / Yihan Zhou / Qi Yang / Yan Gao / Xiuna Yang / Hongzhi Cao / Luke Guddat / Lei Sun / Zihe Rao / Haitao Yang /   Abstract: The entry of coronaviruses is initiated by spike recognition of host cellular receptors, involving proteinaceous and/or glycan receptors. Recently, TMPRSS2 was identified as the proteinaceous ...The entry of coronaviruses is initiated by spike recognition of host cellular receptors, involving proteinaceous and/or glycan receptors. Recently, TMPRSS2 was identified as the proteinaceous receptor for HCoV-HKU1 alongside sialoglycan as a glycan receptor. However, the underlying mechanisms for viral entry remain unknown. Here, we investigated the HCoV-HKU1C spike in the inactive, glycan-activated, and functionally anchored states, revealing that sialoglycan binding induces a conformational change of the NTD and promotes the neighboring RBD of the spike to open for TMPRSS2 recognition, exhibiting a synergistic mechanism for the entry of HCoV-HKU1. The RBD of HCoV-HKU1 features an insertion subdomain that recognizes TMPRSS2 through three previously undiscovered interfaces. Furthermore, structural investigation of HCoV-HKU1A in combination with mutagenesis and binding assays confirms a conserved receptor recognition pattern adopted by HCoV-HKU1. These studies advance our understanding of the complex viral-host interactions during entry, laying the groundwork for developing new therapeutics against coronavirus-associated diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_39036.map.gz emd_39036.map.gz | 216.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-39036-v30.xml emd-39036-v30.xml emd-39036.xml emd-39036.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_39036.png emd_39036.png | 93 KB | ||
| Filedesc metadata |  emd-39036.cif.gz emd-39036.cif.gz | 6.7 KB | ||
| Others |  emd_39036_half_map_1.map.gz emd_39036_half_map_1.map.gz emd_39036_half_map_2.map.gz emd_39036_half_map_2.map.gz | 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-39036 http://ftp.pdbj.org/pub/emdb/structures/EMD-39036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39036 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-39036 | HTTPS FTP |
-Validation report
| Summary document |  emd_39036_validation.pdf.gz emd_39036_validation.pdf.gz | 812.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_39036_full_validation.pdf.gz emd_39036_full_validation.pdf.gz | 811.9 KB | Display | |
| Data in XML |  emd_39036_validation.xml.gz emd_39036_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_39036_validation.cif.gz emd_39036_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39036 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39036 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-39036 | HTTPS FTP |
-Related structure data
| Related structure data |  8y87MC  8y7xC  8y7yC  8y88C  8y89C  8y8aC  8y8bC  8y8cC  8y8dC  8y8eC  8y8fC  8y8gC  8y8hC  8y8iC  8y8jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_39036.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_39036.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_39036_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
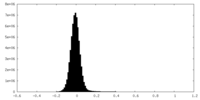
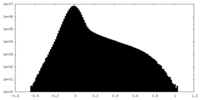
| Density Histograms |
-Half map: #1
| File | emd_39036_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
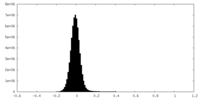
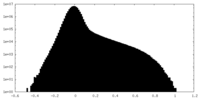
| Density Histograms |
- Sample components
Sample components
-Entire : HKU1B-TMPRSS2 complex
| Entire | Name: HKU1B-TMPRSS2 complex |
|---|---|
| Components |
|
-Supramolecule #1: HKU1B-TMPRSS2 complex
| Supramolecule | Name: HKU1B-TMPRSS2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Human coronavirus HKU1 Human coronavirus HKU1 |
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coronavirus HKU1 (isolate N5) / Strain: isolate N5 Human coronavirus HKU1 (isolate N5) / Strain: isolate N5 |
| Molecular weight | Theoretical: 140.242453 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VIGDFNCTNS FINDYNKTIP RISEDVVDVS LGLGTYYVLN RVYLNTTLLF TGYFPKSGAN FRDLALKGSI YLSTLWYKPP FLSDFNNGI FSKVKNTKLY VNNTLYSEFS TIVIGSVFVN TSYTIVVQPH NGILEITACQ YTMCEYPHTV CKSKGSIRNE S WHIDSSEP ...String: VIGDFNCTNS FINDYNKTIP RISEDVVDVS LGLGTYYVLN RVYLNTTLLF TGYFPKSGAN FRDLALKGSI YLSTLWYKPP FLSDFNNGI FSKVKNTKLY VNNTLYSEFS TIVIGSVFVN TSYTIVVQPH NGILEITACQ YTMCEYPHTV CKSKGSIRNE S WHIDSSEP LCLFKKNFTY NVSADWLYFH FYQERGVFYA YYADVGMPTT FLFSLYLGTI LSHYYVMPLT CNAISSNTDN ET LEYWVTP LSRRQYLLNF DEHGVITNAV DCSSSFLSEI QCKTQSFAPN TGVYDLSGFT VKPVATVYRR IPNLPDCDID NWL NNVSVP SPLNWERRIF SNCNFNLSTL LRLVHVDSFS CNNLDKSKIF GSCFNSITVD KFAIPNRRRD DLQLGSSGFL QSSN YKIDI SSSSCQLYYS LPLVNVTINN FNPSSWNRRY GFGSFNLSSY DVVYSDHCFS VNSDFCPCAD PSVVNSCAKS KPPSA ICPA GTKYRHCDLD TTLYVKNWCR CSCLPDPIST YSPNTCPQKK VVVGIGEHCP GLGINEEKCG TQLNHSSCFC SPDAFL GWS FDSCISNNRC NIFSNFIFNG INSGTTCSND LLYSNTEIST GVCVNYDLYG ITGQGIFKEV SAAYYNNWQN LLYDSNG NI IGFKDFLTNK TYTILPCYSG RVSAAFYQNS SSPALLYRNL KCSYVLNNIS FISQPFYFDS YLGCVLNAVN LTSYSVSS C DLRMGSGFCI DYALPSSGGS GSGISSPYRF VTFEPFNVSF VNDSVETVGG LFEIQIPTNF TIAGHEEFIQ TSSPKVTID CSAFVCSNYA ACHDLLSEYG TFCDNINSIL NEVNDLLDIT QLQVANALMQ GVTLSSNLNT NLHSDVDNID FKSLLGCLGS QCGSSSRSP LEDLLFNKVK LSDVGFVEAY NNCTGGSEIR DLLCVQSFNG IKVLPPILSE TQISGYTTAA TVAAMFPPWS A AAGVPFPL NVQYRINGLG VTMDVLNKNQ KLIANAFNKA LLSIQNGFTA TPSALAKIQS VVNANAQALN SLLQQLFNKF GA ISSSLQE ILSRLDPPEA QVQIDRLING RLTALNAYVS QQLSDITLIK AGASRAIEKV NECVKSQSPR INFCGNGNHI LSL VQNAPY GLLFIHFSYK PTSFKTVLVS PGLCLSGDRG IAPKQGYFIK QNDSWMFTGS SYYYPEPISD KNVVFMNSCS VNFT KAPFI YLNNSIPNLS DFEAELSLWF KNHTSIAPNL TFNSHINATF LDLYYEMNVI QESIKSLN UniProtKB: Spike glycoprotein |
-Macromolecule #2: Transmembrane protease serine 2
| Macromolecule | Name: Transmembrane protease serine 2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: transmembrane protease serine 2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.283629 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSKCSNSGI ECDSSGTCIN PSNWCDGVSH CPGGEDENRC VRLYGPNFIL QVYSSQRKSW HPVCQDDWNE NYGRAACRDM GYKNNFYSS QGIVDDSGST SFMKLNTSAG NVDIYKKLYH SDACSSKAVV SLRCIACGVN LNDDDDKIVG GESALPGAWP W QVSLHVQN ...String: MGSKCSNSGI ECDSSGTCIN PSNWCDGVSH CPGGEDENRC VRLYGPNFIL QVYSSQRKSW HPVCQDDWNE NYGRAACRDM GYKNNFYSS QGIVDDSGST SFMKLNTSAG NVDIYKKLYH SDACSSKAVV SLRCIACGVN LNDDDDKIVG GESALPGAWP W QVSLHVQN VHVCGGSIIT PEWIVTAAHC VEKPLNNPWH WTAFAGILRQ SFMFYGAGYQ VEKVISHPNY DSKTKNNDIA LM KLQKPLT FNDLVKPVCL PNPGMMLQPE QLCWISGWGA TEEKGKTSEV LNAAKVLLIE TQRCNSRYVY DNLITPAMIC AGF LQGNVD SCQGDSGGPL VTSKNNIWWL IGDTSWGSGC AKAYRPGVYG NVMVFTDWIY RQMRADG UniProtKB: Transmembrane protease serine 2 |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 22 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.26 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 89212 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)