+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse BIRC6, Core region | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inhibitor of apoptosis protein / BIRC6 / Smac / APOPTOSIS | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Liu S / Jiang T / Bu F / Zhao J / Wang G / Li N / Gao N / Qiu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy. Authors: Shuo-Shuo Liu / Tian-Xia Jiang / Fan Bu / Ji-Lan Zhao / Guang-Fei Wang / Guo-Heng Yang / Jie-Yan Kong / Yun-Fan Qie / Pei Wen / Li-Bin Fan / Ning-Ning Li / Ning Gao / Xiao-Bo Qiu /  Abstract: Procaspase 9 is the initiator caspase for apoptosis, but how its levels and activities are maintained remains unclear. The gigantic Inhibitor-of-Apoptosis Protein BIRC6/BRUCE/Apollon inhibits both ...Procaspase 9 is the initiator caspase for apoptosis, but how its levels and activities are maintained remains unclear. The gigantic Inhibitor-of-Apoptosis Protein BIRC6/BRUCE/Apollon inhibits both apoptosis and autophagy by promoting ubiquitylation of proapoptotic factors and the key autophagic protein LC3, respectively. Here we show that BIRC6 forms an anti-parallel U-shaped dimer with multiple previously unannotated domains, including a ubiquitin-like domain, and the proapoptotic factor Smac/DIABLO binds BIRC6 in the central cavity. Notably, Smac outcompetes the effector caspase 3 and the pro-apoptotic protease HtrA2, but not procaspase 9, for binding BIRC6 in cells. BIRC6 also binds LC3 through its LC3-interacting region, probably following dimer disruption of this BIRC6 region. Mutation at LC3 ubiquitylation site promotes autophagy and autophagic degradation of BIRC6. Moreover, induction of autophagy promotes autophagic degradation of BIRC6 and caspase 9, but not of other effector caspases. These results are important to understand how the balance between apoptosis and autophagy is regulated under pathophysiological conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38461.map.gz emd_38461.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38461-v30.xml emd-38461-v30.xml emd-38461.xml emd-38461.xml | 12.2 KB 12.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38461.png emd_38461.png | 86.5 KB | ||
| Filedesc metadata |  emd-38461.cif.gz emd-38461.cif.gz | 3.9 KB | ||
| Others |  emd_38461_half_map_1.map.gz emd_38461_half_map_1.map.gz emd_38461_half_map_2.map.gz emd_38461_half_map_2.map.gz | 97.2 MB 97.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38461 http://ftp.pdbj.org/pub/emdb/structures/EMD-38461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38461 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38461 | HTTPS FTP |
-Validation report
| Summary document |  emd_38461_validation.pdf.gz emd_38461_validation.pdf.gz | 687.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38461_full_validation.pdf.gz emd_38461_full_validation.pdf.gz | 687.5 KB | Display | |
| Data in XML |  emd_38461_validation.xml.gz emd_38461_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_38461_validation.cif.gz emd_38461_validation.cif.gz | 16.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38461 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38461 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38461 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38461 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38461.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38461.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||||||||||||||||||
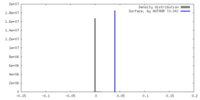
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38461_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38461_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Baculoviral IAP repeat-containing protein 6, Dimer
| Entire | Name: Baculoviral IAP repeat-containing protein 6, Dimer |
|---|---|
| Components |
|
-Supramolecule #1: Baculoviral IAP repeat-containing protein 6, Dimer
| Supramolecule | Name: Baculoviral IAP repeat-containing protein 6, Dimer / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 68.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 154000 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)