+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of prostatic acid phosphatase in human semen | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Acid phosphatase / Acid phosphatase /  CELL CYCLE CELL CYCLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationthiamine phosphate phosphatase activity / positive regulation of adenosine receptor signaling pathway / thiamine metabolic process / Golgi cisterna / adenosine metabolic process /  acid phosphatase / regulation of sensory perception of pain / acid phosphatase / regulation of sensory perception of pain /  lysophosphatidic acid phosphatase activity / lysophosphatidic acid phosphatase activity /  acid phosphatase activity / XMP 5'-nucleosidase activity ...thiamine phosphate phosphatase activity / positive regulation of adenosine receptor signaling pathway / thiamine metabolic process / Golgi cisterna / adenosine metabolic process / acid phosphatase activity / XMP 5'-nucleosidase activity ...thiamine phosphate phosphatase activity / positive regulation of adenosine receptor signaling pathway / thiamine metabolic process / Golgi cisterna / adenosine metabolic process /  acid phosphatase / regulation of sensory perception of pain / acid phosphatase / regulation of sensory perception of pain /  lysophosphatidic acid phosphatase activity / lysophosphatidic acid phosphatase activity /  acid phosphatase activity / XMP 5'-nucleosidase activity / acid phosphatase activity / XMP 5'-nucleosidase activity /  5'-nucleotidase / 5'-nucleotidase /  5'-nucleotidase activity / vesicle membrane / nucleotide metabolic process / 5'-nucleotidase activity / vesicle membrane / nucleotide metabolic process /  choline binding / azurophil granule membrane / lysosome organization / choline binding / azurophil granule membrane / lysosome organization /  phosphatase activity / purine nucleobase metabolic process / phosphatase activity / purine nucleobase metabolic process /  dephosphorylation / dephosphorylation /  multivesicular body / multivesicular body /  protein-tyrosine-phosphatase / protein-tyrosine-phosphatase /  filopodium / filopodium /  protein tyrosine phosphatase activity / lipid metabolic process / apical part of cell / protein tyrosine phosphatase activity / lipid metabolic process / apical part of cell /  lysosome / molecular adaptor activity / lysosomal membrane / Neutrophil degranulation / protein homodimerization activity / lysosome / molecular adaptor activity / lysosomal membrane / Neutrophil degranulation / protein homodimerization activity /  extracellular space / extracellular exosome / identical protein binding / extracellular space / extracellular exosome / identical protein binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
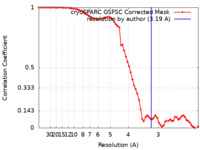
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.19 Å cryo EM / Resolution: 3.19 Å | |||||||||
 Authors Authors | Liu XZ / Li JL / Deng D / Wang X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Biochem Biophys Res Commun / Year: 2024 Journal: Biochem Biophys Res Commun / Year: 2024Title: Purification, identification and Cryo-EM structure of prostatic acid phosphatase in human semen. Authors: Xuanzhong Liu / Lin Yu / Zhili Xia / Jialu Li / Wenbo Meng / Ling Min / Fuping Li / Xiang Wang /  Abstract: Prostatic acid phosphatase (PAP) is a glycoprotein that plays a crucial role in the hydrolysis of phosphate ester present in prostatic exudates. It is a well-established indicator for prostate cancer ...Prostatic acid phosphatase (PAP) is a glycoprotein that plays a crucial role in the hydrolysis of phosphate ester present in prostatic exudates. It is a well-established indicator for prostate cancer due to its elevated serum levels in disease progression. Despite its abundance in semen, PAP's influence on male fertility has not been extensively studied. In our study, we report a significantly optimized method for purifying human endogenous PAP, achieving remarkably high efficiency and active protein recovery rate. This achievement allowed us to better analyze and understand the PAP protein. We determined the cryo-electron microscopic (Cryo-EM) structure of prostatic acid phosphatase in its physiological state for the first time. Our structural and gel filtration analysis confirmed the formation of a tight homodimer structure of human PAP. This functional homodimer displayed an elongated conformation in the cryo-EM structure compared to the previously reported crystal structure. Additionally, there was a notable 5-degree rotation in the angle between the α domain and α/β domain of each monomer. Through structural analysis, we revealed three potential glycosylation sites: Asn94, Asn220, and Asn333. These sites contained varying numbers and forms of glycosyl units, suggesting sugar moieties influence PAP function. Furthermore, we found that the active sites of PAP, His44 and Asp290, are located between the two protein domains. Overall, our study not only provide an optimized approach for PAP purification, but also offer crucial insights into its structural characteristics. These findings lay the groundwork for further investigations into the physiological function and potential therapeutic applications of this important protein. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38393.map.gz emd_38393.map.gz | 28.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38393-v30.xml emd-38393-v30.xml emd-38393.xml emd-38393.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38393_fsc.xml emd_38393_fsc.xml | 6.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_38393.png emd_38393.png | 61.1 KB | ||
| Masks |  emd_38393_msk_1.map emd_38393_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38393.cif.gz emd-38393.cif.gz | 5.4 KB | ||
| Others |  emd_38393_half_map_1.map.gz emd_38393_half_map_1.map.gz emd_38393_half_map_2.map.gz emd_38393_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38393 http://ftp.pdbj.org/pub/emdb/structures/EMD-38393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38393 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38393 | HTTPS FTP |
-Related structure data
| Related structure data |  8xj4MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38393.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38393.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38393_msk_1.map emd_38393_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_38393_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38393_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : structure of prostatic acid phosphatase in human semen
| Entire | Name: structure of prostatic acid phosphatase in human semen |
|---|---|
| Components |
|
-Supramolecule #1: structure of prostatic acid phosphatase in human semen
| Supramolecule | Name: structure of prostatic acid phosphatase in human semen type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Prostatic acid phosphatase
| Macromolecule | Name: Prostatic acid phosphatase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number:  acid phosphatase acid phosphatase |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) / Tissue: Human seminal plasma Homo sapiens (human) / Tissue: Human seminal plasma |
| Molecular weight | Theoretical: 40.262949 KDa |
| Sequence | String: LAKELKFVTL VFRHGDRSPI DTFPTDPIKE SSWPQGFGQL TQLGMEQHYE LGEYIRKRYR KFLNESYKHE QVYIRSTDVD RTLMSAMTN LAALFPPEGV SIWNPILLWQ PIPVHTVPLS EDQLLYLPFR NCPRFQELES ETLKSEEFQK RLHPYKDFIA T LGKLSGLH ...String: LAKELKFVTL VFRHGDRSPI DTFPTDPIKE SSWPQGFGQL TQLGMEQHYE LGEYIRKRYR KFLNESYKHE QVYIRSTDVD RTLMSAMTN LAALFPPEGV SIWNPILLWQ PIPVHTVPLS EDQLLYLPFR NCPRFQELES ETLKSEEFQK RLHPYKDFIA T LGKLSGLH GQDLFGIWSK VYDPLYCESV HNFTLPSWAT EDTMTKLREL SELSLLSLYG IHKQKEKSRL QGGVLVNEIL NH MKRATQI PSYKKLIMYS AHDTTVSGLQ MALDVYNGLL PPYASCHLTE LYFEKGEYFV EMYYRNETQH EPYPLMLPGC SPS CPLERF AELVGPVIPQ DWSTECMTTN S UniProtKB:  Prostatic acid phosphatase Prostatic acid phosphatase |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 4 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #5: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 1 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X