+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C. elegans apo-SID1 structure | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | dsRNA recognition /  MEMBRANE PROTEIN MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA transmembrane transporter activity / dsRNA transport / RNA transport / regulatory ncRNA-mediated post-transcriptional gene silencing /  double-stranded RNA binding / double-stranded RNA binding /  lysosome / lysosome /  membrane / membrane /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) | |||||||||
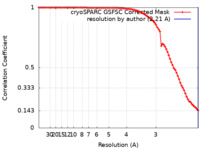
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.21 Å cryo EM / Resolution: 2.21 Å | |||||||||
 Authors Authors | Gong DS | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2024 Journal: Nucleic Acids Res / Year: 2024Title: Structural basis for double-stranded RNA recognition by SID1. Authors: Runhao Wang / Ye Cong / Dandan Qian / Chuangye Yan / Deshun Gong /  Abstract: The nucleic acid transport properties of the systemic RNAi-defective (SID) 1 family make them attractive targets for developing RNA-based therapeutics and drugs. However, the molecular basis for ...The nucleic acid transport properties of the systemic RNAi-defective (SID) 1 family make them attractive targets for developing RNA-based therapeutics and drugs. However, the molecular basis for double-stranded (ds) RNA recognition by SID1 family remains elusive. Here, we report the cryo-EM structures of Caenorhabditis elegans (c) SID1 alone and in complex with dsRNA, both at a resolution of 2.2 Å. The dimeric cSID1 interacts with two dsRNA molecules simultaneously. The dsRNA is located at the interface between β-strand rich domain (BRD)1 and BRD2 and nearly parallel to the membrane plane. In addition to extensive ionic interactions between basic residues and phosphate backbone, several hydrogen bonds are formed between 2'-hydroxyl group of dsRNA and the contact residues. Additionally, the electrostatic potential surface shows three basic regions are fitted perfectly into three major grooves of dsRNA. These structural characteristics enable cSID1 to bind dsRNA in a sequence-independent manner and to distinguish between DNA and RNA. The cSID1 exhibits no conformational changes upon binding dsRNA, with the exception of a few binding surfaces. Structural mapping of dozens of loss-of-function mutations allows potential interpretation of their diverse functional mechanisms. Our study marks an important step toward mechanistic understanding of the SID1 family-mediated dsRNA uptake. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38227.map.gz emd_38227.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38227-v30.xml emd-38227-v30.xml emd-38227.xml emd-38227.xml | 14.7 KB 14.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38227_fsc.xml emd_38227_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_38227.png emd_38227.png | 120.7 KB | ||
| Filedesc metadata |  emd-38227.cif.gz emd-38227.cif.gz | 5.9 KB | ||
| Others |  emd_38227_half_map_1.map.gz emd_38227_half_map_1.map.gz emd_38227_half_map_2.map.gz emd_38227_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38227 http://ftp.pdbj.org/pub/emdb/structures/EMD-38227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38227 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38227 | HTTPS FTP |
-Related structure data
| Related structure data |  8xbsMC  8xc1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38227.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38227.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0825 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_38227_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
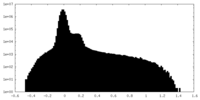
| Projections & Slices |
| ||||||||||||
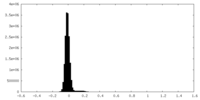
| Density Histograms |
-Half map: #1
| File | emd_38227_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
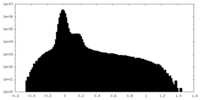
| Projections & Slices |
| ||||||||||||
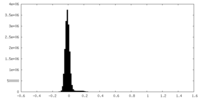
| Density Histograms |
- Sample components
Sample components
-Entire : dimer of sid1
| Entire | Name: dimer of sid1 |
|---|---|
| Components |
|
-Supramolecule #1: dimer of sid1
| Supramolecule | Name: dimer of sid1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) |
-Macromolecule #1: Systemic RNA interference defective protein 1
| Macromolecule | Name: Systemic RNA interference defective protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Caenorhabditis elegans (invertebrata) Caenorhabditis elegans (invertebrata) |
| Molecular weight | Theoretical: 90.081664 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MIRVYLIILM HLVIGLTHHH HHHVEDYKDD DDKQNNSTTP SPIITSSNSS VLVFEISSKM KMIEKKLEAN TVHVLRLELD QSFILDLTK VAAEIVDSSK YSKEDGVILE VTVSNGRDSF LLKLPTVYPN LKLYTDGKLL NPLVEQDFGA HRKRHRIGDP H FHQNLIVT ...String: MIRVYLIILM HLVIGLTHHH HHHVEDYKDD DDKQNNSTTP SPIITSSNSS VLVFEISSKM KMIEKKLEAN TVHVLRLELD QSFILDLTK VAAEIVDSSK YSKEDGVILE VTVSNGRDSF LLKLPTVYPN LKLYTDGKLL NPLVEQDFGA HRKRHRIGDP H FHQNLIVT VQSRLNADID YRLHVTHLDR AQYDFLKFKT GQTTKTLSNQ KLTFVKPIGF FLNCSEQNIS QFHVTLYSED DI CANLITV PANESIYDRS VISDKTHNRR VLSFTKRADI FFTETEISMF KSFRIFVFIA PDDSGCSTNT SRKSFNEKKK ISF EFKKLE NQSYAVPTAL MMIFLTTPCL LFLPIVINII KNSRKLAPSQ SNLISFSPVP SEQRDMDLSH DEQQNTSSEL ENNG EIPAA ENQIVEEITA ENQETSVEEG NREIQVKIPL KQDSLSLHGQ MLQYPVAIIL PVLMHTAIEF HKWTTSTMAN RDEMC FHNH ACARPLGELR AWNNIITNIG YTLYGAIFIV LSICRRGRHE YSHVFGTYEC TLLDVTIGVF MVLQSIASAT YHICPS DVA FQFDTPCIQV ICGLLMVRQW FVRHESPSPA YTNILLVGVV SLNFLISAFS KTSYVRFIIA VIHVIVVGSI CLAKERS LG SEKLKTRFFI MAFSMGNFAA IVMYLTLSAF HLNQIATYCF IINCIMYLMY YGCMKVLHSE RITSKAKLCG ALSLLAWA V AGFFFFQDDT DWTRSAAASR ALNKPCLLLG FFGSHDLWHI FGALAGLFTF IFVSFVDDDL INTRKTSINI F UniProtKB: Systemic RNA interference defective protein 1 |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 4 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #5: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 5 / Number of copies: 2 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm Bright-field microscopy / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.3 µm |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X