+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an anti-phage defense complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DUF4297-HerA / Nuclease / Helicase / DNA BINDING PROTEIN | |||||||||
| Function / homology | : / :  Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
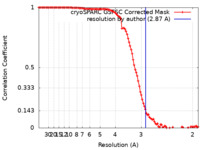
| Method | single particle reconstruction / cryo EM / Resolution: 2.87 Å | |||||||||
 Authors Authors | Wang Y / Deng Z | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Molecular and structural basis of an ATPase-nuclease dual-enzyme anti-phage defense complex. Authors: Qiyin An / Yong Wang / Zhenhua Tian / Jie Han / Jinyue Li / Fumeng Liao / Feiyang Yu / Haiyan Zhao / Yancheng Wen / Heng Zhang / Zengqin Deng /  Abstract: Coupling distinct enzymatic effectors emerges as an efficient strategy for defense against phage infection in bacterial immune responses, such as the widely studied nuclease and cyclase activities in ...Coupling distinct enzymatic effectors emerges as an efficient strategy for defense against phage infection in bacterial immune responses, such as the widely studied nuclease and cyclase activities in the type III CRISPR-Cas system. However, concerted enzymatic activities in other bacterial defense systems are poorly understood. Here, we biochemically and structurally characterize a two-component defense system DUF4297-HerA, demonstrating that DUF4297-HerA confers resistance against phage infection by cooperatively cleaving dsDNA and hydrolyzing ATP. DUF4297 alone forms a dimer, and HerA alone exists as a nonplanar split spiral hexamer, both of which exhibit extremely low enzymatic activity. Interestingly, DUF4297 and HerA assemble into an approximately 1 MDa supramolecular complex, where two layers of DUF4297 (6 DUF4297 molecules per layer) linked via inter-layer dimerization of neighboring DUF4297 molecules are stacked on top of the HerA hexamer. Importantly, the complex assembly promotes dimerization of DUF4297 molecules in the upper layer and enables a transition of HerA from a nonplanar hexamer to a planar hexamer, thus activating their respective enzymatic activities to abrogate phage infection. Together, our findings not only characterize a novel dual-enzyme anti-phage defense system, but also reveal a unique activation mechanism by cooperative complex assembly in bacterial immunity. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38204.map.gz emd_38204.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38204-v30.xml emd-38204-v30.xml emd-38204.xml emd-38204.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_38204_fsc.xml emd_38204_fsc.xml | 10.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_38204.png emd_38204.png | 62.4 KB | ||
| Filedesc metadata |  emd-38204.cif.gz emd-38204.cif.gz | 5.4 KB | ||
| Others |  emd_38204_half_map_1.map.gz emd_38204_half_map_1.map.gz emd_38204_half_map_2.map.gz emd_38204_half_map_2.map.gz | 116 MB 116 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38204 http://ftp.pdbj.org/pub/emdb/structures/EMD-38204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38204 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38204 | HTTPS FTP |
-Validation report
| Summary document |  emd_38204_validation.pdf.gz emd_38204_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38204_full_validation.pdf.gz emd_38204_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_38204_validation.xml.gz emd_38204_validation.xml.gz | 19.1 KB | Display | |
| Data in CIF |  emd_38204_validation.cif.gz emd_38204_validation.cif.gz | 24.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38204 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38204 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38204 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38204 | HTTPS FTP |
-Related structure data
| Related structure data |  8xavMC  8xauC  8xawC  8xaxC  8xayC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38204.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38204.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.95 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38204_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_38204_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DUF4297-HerA
| Entire | Name: DUF4297-HerA |
|---|---|
| Components |
|
-Supramolecule #1: DUF4297-HerA
| Supramolecule | Name: DUF4297-HerA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-binding protein
| Macromolecule | Name: ATP-binding protein / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 64.870047 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSRNNDINAE VVSVSPNKLK ISVDDLEEFK IAEEKLGVGS YLRVSDNQDV ALLAIIDNFS IEVKESQKQK YMIEASPIGL VKNGKFYRG GDSLALPPKK VEPAKLDEII SIYSDSIDIN DRFTFSSLSL NTKVSVPVNG NRFFNKHIAI VGSTGSGKSH T VAKILQKA ...String: MSRNNDINAE VVSVSPNKLK ISVDDLEEFK IAEEKLGVGS YLRVSDNQDV ALLAIIDNFS IEVKESQKQK YMIEASPIGL VKNGKFYRG GDSLALPPKK VEPAKLDEII SIYSDSIDIN DRFTFSSLSL NTKVSVPVNG NRFFNKHIAI VGSTGSGKSH T VAKILQKA VDEKQEGYKG LNNSHIIIFD IHSEYENAFP NSNVLNVDTL TLPYWLLNGD ELEELFLDTE ANDHNQRNVF RQ AITLNKK IHFQGDPATK EIISFHSPYY FDINEVINYI NNRNNERKNK DNEHIWSDEE GNFKFDNENA HRLFKENVTP DGS SAGALN GKLLNFVDRL QSKIFDKRLD FILGEGSKSV TFKETLETLI SYGKDKSNIT ILDVSGVPFE VLSICVSLIS RLIF EFGYH SKKIKRKSNE NQDIPILIVY EEAHKYAPKS DLSKYRTSKE AIERIAKEGR KYGVTLLLAS QRPSEISETI FSQCN TFIS MRLTNPDDQN YVKRLLPDTV GDITNLLPSL KEGEALIMGD SISIPSIVKI EKCTIPPSSI DIKYLDEWRK EWVDSE FDK IIEQWSKS UniProtKB: UNIPROTKB: A0A9X9SUP5 |
-Macromolecule #2: DUF4297
| Macromolecule | Name: DUF4297 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 46.971949 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDRSAVDTIR GYCYQVDKTI IEIFSLPQMD DSIDIECIED VDVYNDGHLT AIQCKYYEST DYNHSVISKP IRLMLSHFKD NKEKGANYY LYGHYKSGQE KLTLPLKVDF FKSNFLTYTE KKIKHEYHIE NGLTEEDLQA FLDRLVININ AKSFDDQKKE T IQIIKNHF ...String: MDRSAVDTIR GYCYQVDKTI IEIFSLPQMD DSIDIECIED VDVYNDGHLT AIQCKYYEST DYNHSVISKP IRLMLSHFKD NKEKGANYY LYGHYKSGQE KLTLPLKVDF FKSNFLTYTE KKIKHEYHIE NGLTEEDLQA FLDRLVININ AKSFDDQKKE T IQIIKNHF QCEDYEAEHY LYSNAFRKTY DISCNKKDRR IKKSDFVESI NKSKVLFNIW FYQYEGRKEY LRKLKESFIR RS VNTSPYA RFFILEFQDK TDIKTVKDCI YKIQSNWSNL SKRTDRPYSP FLLFHGTSDA NLYELKNQLF NEDLIFTDGY PFK GSVFTP KMLIEGFSNK EIHFQFINDI DDFNETLNSI NIRKEVYQFY TENCLDIPSQ LPQVNIQVKD FADIKEIV UniProtKB: UNIPROTKB: A0A9X9SUN3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X