+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human SLC15A3 (dimer) | |||||||||
 Map data Map data | B-factor sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidoglycan transmembrane transporter activity / peptidoglycan transport / Proton/oligopeptide cotransporters / dipeptide import across plasma membrane / peptide:proton symporter activity / dipeptide transmembrane transporter activity / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / monoatomic ion transport / protein transport / endosome membrane ...peptidoglycan transmembrane transporter activity / peptidoglycan transport / Proton/oligopeptide cotransporters / dipeptide import across plasma membrane / peptide:proton symporter activity / dipeptide transmembrane transporter activity / positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway / monoatomic ion transport / protein transport / endosome membrane / lysosomal membrane / intracellular membrane-bounded organelle / innate immune response Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.44 Å | |||||||||
 Authors Authors | Kasai S / Zhang Z / Ohto U / Shimizu T | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of human SLC15A3 (outward-facing partially occluded) Authors: Zhang Z / Kasai S / Sakaniwa K / Fujimura A / Ohto U / Shimizu T | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37898.map.gz emd_37898.map.gz | 46.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37898-v30.xml emd-37898-v30.xml emd-37898.xml emd-37898.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37898.png emd_37898.png | 80.9 KB | ||
| Filedesc metadata |  emd-37898.cif.gz emd-37898.cif.gz | 5.3 KB | ||
| Others |  emd_37898_additional_1.map.gz emd_37898_additional_1.map.gz emd_37898_half_map_1.map.gz emd_37898_half_map_1.map.gz emd_37898_half_map_2.map.gz emd_37898_half_map_2.map.gz | 45.4 MB 86 MB 86 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37898 http://ftp.pdbj.org/pub/emdb/structures/EMD-37898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37898 | HTTPS FTP |
-Validation report
| Summary document |  emd_37898_validation.pdf.gz emd_37898_validation.pdf.gz | 867.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37898_full_validation.pdf.gz emd_37898_full_validation.pdf.gz | 866.6 KB | Display | |
| Data in XML |  emd_37898_validation.xml.gz emd_37898_validation.xml.gz | 13.3 KB | Display | |
| Data in CIF |  emd_37898_validation.cif.gz emd_37898_validation.cif.gz | 15.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37898 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37898 | HTTPS FTP |
-Related structure data
| Related structure data |  8wx2MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37898.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37898.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | B-factor sharpened map | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
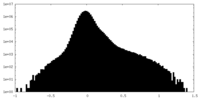
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: Unsharpened map
| File | emd_37898_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
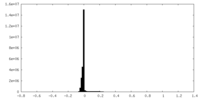
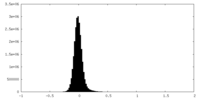
| Density Histograms |
-Half map: #2
| File | emd_37898_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
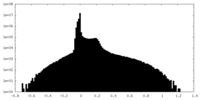
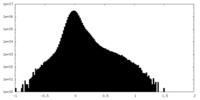
| Density Histograms |
-Half map: #1
| File | emd_37898_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
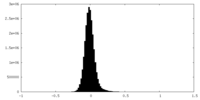
| Density Histograms |
- Sample components
Sample components
-Entire : Human SLC15A3
| Entire | Name: Human SLC15A3 |
|---|---|
| Components |
|
-Supramolecule #1: Human SLC15A3
| Supramolecule | Name: Human SLC15A3 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Solute carrier family 15 member 3
| Macromolecule | Name: Solute carrier family 15 member 3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 67.140953 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSYYHHHHHH DYKDDDDKLE VLFQGPEFMP APRAREQPRV PGERQPLLPR GARGPRRWRR AAGAAVLLVE MLERAAFFGV TANLVLYLN STNFNWTGEQ ATRAALVFLG ASYLLAPVGG WLADVYLGRY RAVALSLLLY LAASGLLPAT AFPDGRSSFC G EMPASPLG ...String: MSYYHHHHHH DYKDDDDKLE VLFQGPEFMP APRAREQPRV PGERQPLLPR GARGPRRWRR AAGAAVLLVE MLERAAFFGV TANLVLYLN STNFNWTGEQ ATRAALVFLG ASYLLAPVGG WLADVYLGRY RAVALSLLLY LAASGLLPAT AFPDGRSSFC G EMPASPLG PACPSAGCPR SSPSPYCAPV LYAGLLLLGL AASSVRSNLT SFGADQVMDL GRDATRRFFN WFYWSINLGA VL SLLVVAF IQQNISFLLG YSIPVGCVGL AFFIFLFATP VFITKPPMGS QVSSMLKLAL QNCCPQLWQR HSARDRQCAR VLA DERSPQ PGASPQEDIA NFQVLVKILP VMVTLVPYWM VYFQMQSTYV LQGLHLHIPN IFPANPANIS VALRAQGSSY TIPE AWLLL ANVVVVLILV PLKDRLIDPL LLRCKLLPSA LQKMALGMFF GFTSVIVAGV LEMERLHYIH HNETVSQQIG EVLYN AAPL SIWWQIPQYL LIGISEIFAS IPGLEFAYSE APRSMQGAIM GIFFCLSGVG SLLGSSLVAL LSLPGGWLHC PKDFGN INN CRMDLYFFLL AGIQAVTALL FVWIAGRYER ASQGPASHSR FSRDRG UniProtKB: Solute carrier family 15 member 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.44 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 91199 |
| Initial angle assignment | Type: NOT APPLICABLE |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller







 Z
Z Y
Y X
X