[English] 日本語
 Yorodumi
Yorodumi- EMDB-37619: Cryo-EM structure of the proximal rod-export apparatus and FlgF w... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CCW state | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Flagellum / Flagellar motor / Proximal rod / Export apparatus / FlgF / MOTOR PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationbacterial-type flagellum basal body, rod / bacterial-type flagellum organization / bacterial-type flagellum basal body, MS ring / bacterial-type flagellum basal body / bacterial-type flagellum-dependent swarming motility / bacterial-type flagellum assembly / cytoskeletal motor activity / bacterial-type flagellum-dependent cell motility / protein secretion / protein targeting ...bacterial-type flagellum basal body, rod / bacterial-type flagellum organization / bacterial-type flagellum basal body, MS ring / bacterial-type flagellum basal body / bacterial-type flagellum-dependent swarming motility / bacterial-type flagellum assembly / cytoskeletal motor activity / bacterial-type flagellum-dependent cell motility / protein secretion / protein targeting / structural molecule activity / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) | ||||||||||||
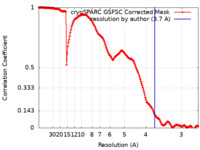
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Tan JX / Zhang L / Zhou Y / Zhu YQ | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of the proximal rod-export apparatus and FlgF within the motor-hook complex in the CCW state Authors: Tan JX / Zhang L / Zhou Y / Zhu YQ | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37619.map.gz emd_37619.map.gz | 483.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37619-v30.xml emd-37619-v30.xml emd-37619.xml emd-37619.xml | 24.4 KB 24.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37619_fsc.xml emd_37619_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_37619.png emd_37619.png | 67.1 KB | ||
| Filedesc metadata |  emd-37619.cif.gz emd-37619.cif.gz | 7 KB | ||
| Others |  emd_37619_half_map_1.map.gz emd_37619_half_map_1.map.gz emd_37619_half_map_2.map.gz emd_37619_half_map_2.map.gz | 475.6 MB 475.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37619 http://ftp.pdbj.org/pub/emdb/structures/EMD-37619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37619 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37619 | HTTPS FTP |
-Validation report
| Summary document |  emd_37619_validation.pdf.gz emd_37619_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37619_full_validation.pdf.gz emd_37619_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_37619_validation.xml.gz emd_37619_validation.xml.gz | 26.3 KB | Display | |
| Data in CIF |  emd_37619_validation.cif.gz emd_37619_validation.cif.gz | 34.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37619 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37619 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37619 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37619 | HTTPS FTP |
-Related structure data
| Related structure data |  8wlhMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37619.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37619.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.332 Å | ||||||||||||||||||||||||||||||||||||
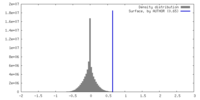
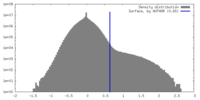
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37619_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
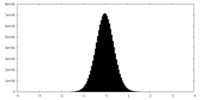
| Density Histograms |
-Half map: #2
| File | emd_37619_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
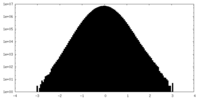
| Density Histograms |
- Sample components
Sample components
-Entire : C ring-containing flagellar motor-hook complex in the CCW state
| Entire | Name: C ring-containing flagellar motor-hook complex in the CCW state |
|---|---|
| Components |
|
-Supramolecule #1: C ring-containing flagellar motor-hook complex in the CCW state
| Supramolecule | Name: C ring-containing flagellar motor-hook complex in the CCW state type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
-Macromolecule #1: Flagellar biosynthetic protein FliQ
| Macromolecule | Name: Flagellar biosynthetic protein FliQ / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 9.606758 KDa |
| Sequence | String: MTPESVMMMG TEAMKVALAL AAPLLLVALI TGLIISILQA ATQINEMTLS FIPKIVAVFI AIIVAGPWML NLLLDYVRTL FSNLPYIIG UniProtKB: Flagellar biosynthetic protein FliQ |
-Macromolecule #2: Flagellar biosynthetic protein FliR
| Macromolecule | Name: Flagellar biosynthetic protein FliR / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 28.938865 KDa |
| Sequence | String: MIQVTSEQWL YWLHLYFWPL LRVLALISTA PILSERAIPK RVKLGLGIMI TLVIAPSLPA NDTPLFSIAA LWLAMQQILI GIALGFTMQ FAFAAVRTAG EFIGLQMGLS FATFVDPGSH LNMPVLARIM DMLAMLLFLT FNGHLWLISL LVDTFHTLPI G SNPVNSNA ...String: MIQVTSEQWL YWLHLYFWPL LRVLALISTA PILSERAIPK RVKLGLGIMI TLVIAPSLPA NDTPLFSIAA LWLAMQQILI GIALGFTMQ FAFAAVRTAG EFIGLQMGLS FATFVDPGSH LNMPVLARIM DMLAMLLFLT FNGHLWLISL LVDTFHTLPI G SNPVNSNA FMALARAGGL IFLNGLMLAL PVITLLLTLN LALGLLNRMA PQLSIFVIGF PLTLTVGIML MAALMPLIAP FC EHLFSEI FNLLADIVSE MPINNNP UniProtKB: Flagellar biosynthetic protein FliR |
-Macromolecule #3: Flagellar biosynthetic protein FliP
| Macromolecule | Name: Flagellar biosynthetic protein FliP / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 26.801086 KDa |
| Sequence | String: MRRLLFLSLA GLWLFSPAAA AQLPGLISQP LAGGGQSWSL SVQTLVFITS LTFLPAILLM MTSFTRIIIV FGLLRNALGT PSAPPNQVL LGLALFLTFF IMSPVIDKIY VDAYQPFSEQ KISMQEALDK GAQPLRAFML RQTREADLAL FARLANSGPL Q GPEAVPMR ...String: MRRLLFLSLA GLWLFSPAAA AQLPGLISQP LAGGGQSWSL SVQTLVFITS LTFLPAILLM MTSFTRIIIV FGLLRNALGT PSAPPNQVL LGLALFLTFF IMSPVIDKIY VDAYQPFSEQ KISMQEALDK GAQPLRAFML RQTREADLAL FARLANSGPL Q GPEAVPMR ILLPAYVTSE LKTAFQIGFT IFIPFLIIDL VIASVLMALG MMMVPPATIA LPFKLMLFVL VDGWQLLMGS LA QSFYS UniProtKB: Flagellar biosynthetic protein FliP |
-Macromolecule #4: Flagellar hook-basal body complex protein FliE
| Macromolecule | Name: Flagellar hook-basal body complex protein FliE / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 11.087662 KDa |
| Sequence | String: MAAIQGIEGV ISQLQATAMA ARGQDTHSQS TVSFAGQLHA ALDRISDRQA AARVQAEKFT LGEPGIALND VMADMQKASV SMQMGIQVR NKLVAAYQEV MSMQV UniProtKB: Flagellar hook-basal body complex protein FliE |
-Macromolecule #5: Flagellar basal body rod protein FlgB
| Macromolecule | Name: Flagellar basal body rod protein FlgB / type: protein_or_peptide / ID: 5 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 15.145061 KDa |
| Sequence | String: MLDRLDAALR FQQEALNLRA QRQEILAANI ANADTPGYQA RDIDFASELK KVMVRGREET GGVALTLTSS HHIPAQAVSS PAVDLLYRV PDQPSLDGNT VDMDRERTQF ADNSLKYQMG LTVLGSQLKG MMNVLQGGN UniProtKB: Flagellar basal body rod protein FlgB |
-Macromolecule #6: Flagellar basal-body rod protein FlgC
| Macromolecule | Name: Flagellar basal-body rod protein FlgC / type: protein_or_peptide / ID: 6 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 13.991889 KDa |
| Sequence | String: MALLNIFDIA GSALAAQSKR LNVAASNLAN ADSVTGPDGQ PYRAKQVVFQ VDAAPGQATG GVKVASVIES QAPEKLVYEP GNPLADANG YVKMPNVDVV GEMVNTMSAS RSYQANIEVL NTVKSMMLKT LTLGQ UniProtKB: Flagellar basal-body rod protein FlgC |
-Macromolecule #7: Flagellar M-ring protein
| Macromolecule | Name: Flagellar M-ring protein / type: protein_or_peptide / ID: 7 / Number of copies: 11 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 61.295645 KDa |
| Sequence | String: MSATASTATQ PKPLEWLNRL RANPRIPLIV AGSAAVAIVV AMVLWAKTPD YRTLFSNLSD QDGGAIVAQL TQMNIPYRFA NGSGAIEVP ADKVHELRLR LAQQGLPKGG AVGFELLDQE KFGISQFSEQ VNYQRALEGE LARTIETLGP VKSARVHLAM P KPSLFVRE ...String: MSATASTATQ PKPLEWLNRL RANPRIPLIV AGSAAVAIVV AMVLWAKTPD YRTLFSNLSD QDGGAIVAQL TQMNIPYRFA NGSGAIEVP ADKVHELRLR LAQQGLPKGG AVGFELLDQE KFGISQFSEQ VNYQRALEGE LARTIETLGP VKSARVHLAM P KPSLFVRE QKSPSASVTV TLEPGRALDE GQISAVVHLV SSAVAGLPPG NVTLVDQSGH LLTQSNTSGR DLNDAQLKFA ND VESRIQR RIEAILSPIV GNGNVHAQVT AQLDFANKEQ TEEHYSPNGD ASKATLRSRQ LNISEQVGAG YPGGVPGALS NQP APPNEA PIATPPTNQQ NAQNTPQTST STNSNSAGPR STQRNETSNY EVDRTIRHTK MNVGDIERLS VAVVVNYKTL ADGK PLPLT ADQMKQIEDL TREAMGFSDK RGDTLNVVNS PFSAVDNTGG ELPFWQQQSF IDQLLAAGRW LLVLVVAWIL WRKAV RPQL TRRVEEAKAA QEQAQVRQET EEAVEVRLSK DEQLQQRRAN QRLGAEVMSQ RIREMSDNDP RVVALVIRQW MSNDHE UniProtKB: Flagellar M-ring protein |
-Macromolecule #8: Flagellar basal-body rod protein FlgF
| Macromolecule | Name: Flagellar basal-body rod protein FlgF / type: protein_or_peptide / ID: 8 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 (bacteria) |
| Molecular weight | Theoretical: 26.121223 KDa |
| Sequence | String: MDHAIYTAMG AASQTLNQQA VTASNLANAS TPGFRAQLNA LRAVPVDGLS LATRTLVTAS TPGADMTPGQ LDYTSRPLDV ALQQDGWLV VQAADGAEGY TRNGNIQVGP TGQLTIQGHP VIGEGGPITV PEGSEITIAA DGTISALNPG DPPNTVAPVG R LKLVKAEG ...String: MDHAIYTAMG AASQTLNQQA VTASNLANAS TPGFRAQLNA LRAVPVDGLS LATRTLVTAS TPGADMTPGQ LDYTSRPLDV ALQQDGWLV VQAADGAEGY TRNGNIQVGP TGQLTIQGHP VIGEGGPITV PEGSEITIAA DGTISALNPG DPPNTVAPVG R LKLVKAEG NEVQRSDDGL FRLTAEAQAE RGAVLAADPS IRIMSGVLEG SNVKPVEAMT DMIANARRFE MQMKVITSVD EN EGRANQL LSMS UniProtKB: Flagellar basal-body rod protein FlgF |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Support film - Film thickness: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. / Pretreatment - Atmosphere: AIR | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: Blot time: 2 Blot force: -15 Wait time: 60 Blot total: 1. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: Other / Chain - Initial model type: in silico model / Details: Model-Angelo |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8wlh: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)