+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a bacterial protein | |||||||||
 Map data Map data | main map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | bacterial immune proteins / IMMUNE SYSTEM | |||||||||
| Biological species |  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) | |||||||||
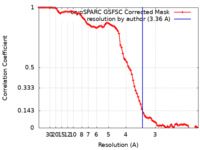
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Yu G / Liao F / Li X / Li Z / Zhang H | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of a bacterial protein Authors: Yu G / Liao F / Li X / Li Z / Zhang H | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37598.map.gz emd_37598.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37598-v30.xml emd-37598-v30.xml emd-37598.xml emd-37598.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37598_fsc.xml emd_37598_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_37598.png emd_37598.png | 177.7 KB | ||
| Filedesc metadata |  emd-37598.cif.gz emd-37598.cif.gz | 4.8 KB | ||
| Others |  emd_37598_half_map_1.map.gz emd_37598_half_map_1.map.gz emd_37598_half_map_2.map.gz emd_37598_half_map_2.map.gz | 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37598 http://ftp.pdbj.org/pub/emdb/structures/EMD-37598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37598 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37598 | HTTPS FTP |
-Validation report
| Summary document |  emd_37598_validation.pdf.gz emd_37598_validation.pdf.gz | 942.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37598_full_validation.pdf.gz emd_37598_full_validation.pdf.gz | 942.2 KB | Display | |
| Data in XML |  emd_37598_validation.xml.gz emd_37598_validation.xml.gz | 16.3 KB | Display | |
| Data in CIF |  emd_37598_validation.cif.gz emd_37598_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37598 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37598 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37598 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37598 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37598.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37598.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07578 Å | ||||||||||||||||||||||||||||||||||||
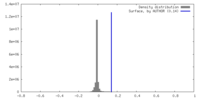
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_37598_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
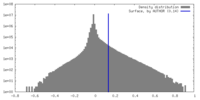
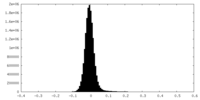
| Density Histograms |
-Half map: half map
| File | emd_37598_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
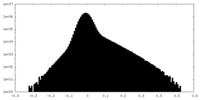
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of a bacterial protein
| Entire | Name: Cryo-EM structure of a bacterial protein |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of a bacterial protein
| Supramolecule | Name: Cryo-EM structure of a bacterial protein / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) |
| Molecular weight | Theoretical: 400 KDa |
-Macromolecule #1: bacterial protein
| Macromolecule | Name: bacterial protein / type: protein_or_peptide / ID: 1 / Details: AMPPNP, MG / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Paenibacillus sp. 453mf (bacteria) Paenibacillus sp. 453mf (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MIGVNRMTEA STYIGTVQDV NGANIRVVLD INTISSLKFV DGQGYRIGQI GSFVRIPIGY INLFGIVSQV GAGAVPDKLL EVEPYGHRWI SVQLVGEEGI KKEFERGVSQ YPTIGDKVHI VTEPDLKKIY GTQNKKYISL GNIASVDSIP ALVNIDTLVT RHSAVLGSTG ...String: MIGVNRMTEA STYIGTVQDV NGANIRVVLD INTISSLKFV DGQGYRIGQI GSFVRIPIGY INLFGIVSQV GAGAVPDKLL EVEPYGHRWI SVQLVGEEGI KKEFERGVSQ YPTIGDKVHI VTEPDLKKIY GTQNKKYISL GNIASVDSIP ALVNIDTLVT RHSAVLGSTG SGKSTTVTSI LQRISDMSQF PSARIIVFDI HGEYAAAFKG KAKVYKVTPS NNELKLSIPY WALTCDEFLS VAFGGLEGSG RNALIDKIYE LKLQTLKRQE YEGINEDSLT VDTPIPFSIH KLWFDLYRAE ISTHYVQGSH SEENEALLLG EDGNPVQKGD SLKVVPPIYM PHTQAQGATK IYLSNRGKNI RKPLEGLASL LKDPRYEFLF NADDWSVNLD GKTNKDLDAL LETWVGSEES ISIFDLSGMP SSILDTLIGI LIRILYDSLF WSRNQPEGGR ERPLLVVLEE AHTYLGKDSR GIAIDGVRKI VKEGRKYGIG MMLVSQRPSE IDSTILSQCG TLFALRMNNS SDRNHVLGAV SDSFEGLMGM LPTLRTGEAI IIGESVRLPM RTIISPPPFG RRPDSLDPDV TAKWSNNRVQ GDYKEVLTLW RQKKVRSQRI VENIKRLPVV NEGEMTDMVR EMVTSSNILS IGYEADSMTL EIEFNHGLVY QYYDVPETLH TELLAAESHG KFFNSQIKNN YRFSRI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)