+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Prefusion RSV F Bound to Lonafarnib and D25 Fab | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Respiratory syncytial virus / F protein / Respiratory syncytial virus / F protein /  Lonafarnib / Lonafarnib /  VIRAL PROTEIN/IMMUNE SYSTEM / VIRAL PROTEIN/IMMUNE SYSTEM /  VIRAL PROTEIN-IMMUNE SYSTEM complex VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of syncytium formation by virus / Assembly and release of respiratory syncytial virus (RSV) virions / Translation of respiratory syncytial virus mRNAs / Respiratory syncytial virus (RSV) attachment and entry / RSV-host interactions / Maturation of hRSV A proteins / host cell Golgi membrane /  virion component / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell ...positive regulation of syncytium formation by virus / Assembly and release of respiratory syncytial virus (RSV) virions / Translation of respiratory syncytial virus mRNAs / Respiratory syncytial virus (RSV) attachment and entry / RSV-host interactions / Maturation of hRSV A proteins / host cell Golgi membrane / virion component / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell ...positive regulation of syncytium formation by virus / Assembly and release of respiratory syncytial virus (RSV) virions / Translation of respiratory syncytial virus mRNAs / Respiratory syncytial virus (RSV) attachment and entry / RSV-host interactions / Maturation of hRSV A proteins / host cell Golgi membrane /  virion component / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell / fusion of virus membrane with host plasma membrane / virion component / entry receptor-mediated virion attachment to host cell / symbiont entry into host cell / fusion of virus membrane with host plasma membrane /  viral envelope / host cell plasma membrane / virion membrane / identical protein binding / viral envelope / host cell plasma membrane / virion membrane / identical protein binding /  plasma membrane plasma membraneSimilarity search - Function | |||||||||
| Biological species |   Human respiratory syncytial virus A (strain A2) / Human respiratory syncytial virus A (strain A2) /   Enterobacteria phage T4 (virus) / Enterobacteria phage T4 (virus) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.17 Å cryo EM / Resolution: 3.17 Å | |||||||||
 Authors Authors | Yang Q / Xue B / Liu F / Peng W / Chen X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Signal Transduct Target Ther / Year: 2024 Journal: Signal Transduct Target Ther / Year: 2024Title: Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein. Authors: Qi Yang / Bao Xue / Fengjiang Liu / Yongzhi Lu / Jielin Tang / Mengrong Yan / Qiong Wu / Ruyi Chen / Anqi Zhou / Lijie Liu / Junjun Liu / Changbin Qu / Qingxin Wu / Muqing Fu / Jiayi Zhong / ...Authors: Qi Yang / Bao Xue / Fengjiang Liu / Yongzhi Lu / Jielin Tang / Mengrong Yan / Qiong Wu / Ruyi Chen / Anqi Zhou / Lijie Liu / Junjun Liu / Changbin Qu / Qingxin Wu / Muqing Fu / Jiayi Zhong / Jianwei Dong / Sijie Chen / Fan Wang / Yuan Zhou / Jie Zheng / Wei Peng / Jinsai Shang / Xinwen Chen /  Abstract: Respiratory syncytial virus (RSV) is the major cause of bronchiolitis and pneumonia in young children and the elderly. There are currently no approved RSV-specific therapeutic small molecules ...Respiratory syncytial virus (RSV) is the major cause of bronchiolitis and pneumonia in young children and the elderly. There are currently no approved RSV-specific therapeutic small molecules available. Using high-throughput antiviral screening, we identified an oral drug, the prenylation inhibitor lonafarnib, which showed potent inhibition of the RSV fusion process. Lonafarnib exhibited antiviral activity against both the RSV A and B genotypes and showed low cytotoxicity in HEp-2 and human primary bronchial epithelial cells (HBEC). Time-of-addition and pseudovirus assays demonstrated that lonafarnib inhibits RSV entry, but has farnesyltransferase-independent antiviral efficacy. Cryo-electron microscopy revealed that lonafarnib binds to a triple-symmetric pocket within the central cavity of the RSV F metastable pre-fusion conformation. Mutants at the RSV F sites interacting with lonafarnib showed resistance to lonafarnib but remained fully sensitive to the neutralizing monoclonal antibody palivizumab. Furthermore, lonafarnib dose-dependently reduced the replication of RSV in BALB/c mice. Collectively, lonafarnib could be a potential fusion inhibitor for RSV infection. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37210.map.gz emd_37210.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37210-v30.xml emd-37210-v30.xml emd-37210.xml emd-37210.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37210.png emd_37210.png | 20.2 KB | ||
| Filedesc metadata |  emd-37210.cif.gz emd-37210.cif.gz | 6.1 KB | ||
| Others |  emd_37210_half_map_1.map.gz emd_37210_half_map_1.map.gz emd_37210_half_map_2.map.gz emd_37210_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37210 http://ftp.pdbj.org/pub/emdb/structures/EMD-37210 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37210 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37210 | HTTPS FTP |
-Related structure data
| Related structure data |  8kg5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37210.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37210.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.73 Å | ||||||||||||||||||||||||||||||||||||
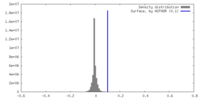
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37210_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
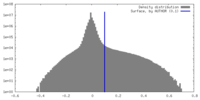
| Density Histograms |
-Half map: #1
| File | emd_37210_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
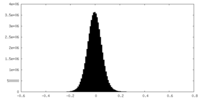
| Density Histograms |
- Sample components
Sample components
-Entire : Prefusion RSV F Bound to Lonafarnib and D25 Fab
| Entire | Name: Prefusion RSV F Bound to Lonafarnib and D25 Fab |
|---|---|
| Components |
|
-Supramolecule #1: Prefusion RSV F Bound to Lonafarnib and D25 Fab
| Supramolecule | Name: Prefusion RSV F Bound to Lonafarnib and D25 Fab / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   Human respiratory syncytial virus A (strain A2) Human respiratory syncytial virus A (strain A2) |
-Macromolecule #1: Fusion glycoprotein F0,Fibritin
| Macromolecule | Name: Fusion glycoprotein F0,Fibritin / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
| Molecular weight | Theoretical: 64.530602 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MELLILKANA ITTILTAVTF CFASGQNITE EFYQSTCSAV SKGYLSALRT GWYTSVITIE LSNIKENKCN GTDAKVKLIK QELDKYKNA VTELQLLMQS TPATNNRARR ELPRFMNYTL NNAKKTNVTL SKKRKRRFLG FLLGVGSAIA SGVAVCKVLH L EGEVNKIK ...String: MELLILKANA ITTILTAVTF CFASGQNITE EFYQSTCSAV SKGYLSALRT GWYTSVITIE LSNIKENKCN GTDAKVKLIK QELDKYKNA VTELQLLMQS TPATNNRARR ELPRFMNYTL NNAKKTNVTL SKKRKRRFLG FLLGVGSAIA SGVAVCKVLH L EGEVNKIK SALLSTNKAV VSLSNGVSVL TFKVLDLKNY IDKQLLPILN KQSCSISNIE TVIEFQQKNN RLLEITREFS VN AGVTTPV STYMLTNSEL LSLINDMPIT NDQKKLMSNN VQIVRQQSYS IMCIIKEEVL AYVVQLPLYG VIDTPCWKLH TSP LCTTNT KEGSNICLTR TDRGWYCDNA GSVSFFPQAE TCKVQSNRVF CDTMNSLTLP SEVNLCNVDI FNPKYDCKIM TSKT DVSSS VITSLGAIVS CYGKTKCTAS NKNRGIIKTF SNGCDYVSNK GVDTVSVGNT LYYVNKQEGK SLYVKGEPII NFYDP LVFP SDEFDASISQ VNEKINQSLA FIRKSDELLG SGYIPEAPRD GQAYVRKDGE WVFLSTFLSG LEVLFQGPGG WSHPQF EKG GGSGGGSGGS AWSHPQFEKG GS UniProtKB: Fusion glycoprotein F0, Fibritin |
-Macromolecule #2: D25 heavy chain
| Macromolecule | Name: D25 heavy chain / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 24.504629 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGSSVMV SCQASGGPLR NYIINWLRQA PGQGPEWMGG IIPVLGTVHY APKFQGRVTI TADESTDTAY IHLISLRSE DTAMYYCATE TALVVSTTYL PHYFDNWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN ...String: QVQLVQSGAE VKKPGSSVMV SCQASGGPLR NYIINWLRQA PGQGPEWMGG IIPVLGTVHY APKFQGRVTI TADESTDTAY IHLISLRSE DTAMYYCATE TALVVSTTYL PHYFDNWGQG TLVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF P EPVTVSWN SGALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDK |
-Macromolecule #3: D25 light chain
| Macromolecule | Name: D25 light chain / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.325865 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIQMTQSPSS LSAAVGDRVT ITCQASQDIV NYLNWYQQKP GKAPKLLIYV ASNLETGVPS RFSGSGSGTD FSLTISSLQP EDVATYYCQ QYDNLPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS ...String: DIQMTQSPSS LSAAVGDRVT ITCQASQDIV NYLNWYQQKP GKAPKLLIYV ASNLETGVPS RFSGSGSGTD FSLTISSLQP EDVATYYCQ QYDNLPLTFG GGTKVEIKRT VAAPSVFIFP PSDEQLKSGT ASVVCLLNNF YPREAKVQWK VDNALQSGNS Q ESVTEQDS KDSTYSLSST LTLSKADYEK HKVYACEVTH QGLSSPVTKS FNRGEC |
-Macromolecule #4: 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEP...
| Macromolecule | Name: 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE type: ligand / ID: 4 / Number of copies: 3 / Formula: 336 |
|---|---|
| Molecular weight | Theoretical: 638.822 Da |
| Chemical component information | 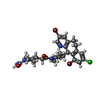 ChemComp-336: |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm Bright-field microscopy / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.17 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 37059 |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)