[English] 日本語
 Yorodumi
Yorodumi- EMDB-36800: Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state | |||||||||
 Map data Map data | main map for KtrAB | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | KtrAB / RCK / potassium / transporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpotassium:chloride symporter activity / monoatomic cation transmembrane transporter activity / potassium ion transport / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
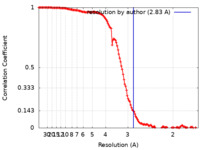
| Method | single particle reconstruction / cryo EM / Resolution: 2.83 Å | |||||||||
 Authors Authors | Chang YK / Chiang WT / Hu NJ / Tsai MD | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural basis and synergism of ATP and Na activation in bacterial K uptake system KtrAB. Authors: Wesley Tien Chiang / Yao-Kai Chang / Wei-Han Hui / Shu-Wei Chang / Chen-Yi Liao / Yi-Chuan Chang / Chun-Jung Chen / Wei-Chen Wang / Chien-Chen Lai / Chun-Hsiung Wang / Siou-Ying Luo / Ya- ...Authors: Wesley Tien Chiang / Yao-Kai Chang / Wei-Han Hui / Shu-Wei Chang / Chen-Yi Liao / Yi-Chuan Chang / Chun-Jung Chen / Wei-Chen Wang / Chien-Chen Lai / Chun-Hsiung Wang / Siou-Ying Luo / Ya-Ping Huang / Shan-Ho Chou / Tzyy-Leng Horng / Ming-Hon Hou / Stephen P Muench / Ren-Shiang Chen / Ming-Daw Tsai / Nien-Jen Hu /   Abstract: The K uptake system KtrAB is essential for bacterial survival in low K environments. The activity of KtrAB is regulated by nucleotides and Na. Previous studies proposed a putative gating mechanism of ...The K uptake system KtrAB is essential for bacterial survival in low K environments. The activity of KtrAB is regulated by nucleotides and Na. Previous studies proposed a putative gating mechanism of KtrB regulated by KtrA upon binding to ATP or ADP. However, how Na activates KtrAB and the Na binding site remain unknown. Here we present the cryo-EM structures of ATP- and ADP-bound KtrAB from Bacillus subtilis (BsKtrAB) both solved at 2.8 Å. A cryo-EM density at the intra-dimer interface of ATP-KtrA was identified as Na, as supported by X-ray crystallography and ICP-MS. Thermostability assays and functional studies demonstrated that Na binding stabilizes the ATP-bound BsKtrAB complex and enhances its K flux activity. Comparing ATP- and ADP-BsKtrAB structures suggests that BsKtrB Arg417 and Phe91 serve as a channel gate. The synergism of ATP and Na in activating BsKtrAB is likely applicable to Na-activated K channels in central nervous system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36800.map.gz emd_36800.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36800-v30.xml emd-36800-v30.xml emd-36800.xml emd-36800.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36800_fsc.xml emd_36800_fsc.xml | 13.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_36800.png emd_36800.png | 126.9 KB | ||
| Masks |  emd_36800_msk_1.map emd_36800_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-36800.cif.gz emd-36800.cif.gz | 6.7 KB | ||
| Others |  emd_36800_additional_1.map.gz emd_36800_additional_1.map.gz emd_36800_additional_2.map.gz emd_36800_additional_2.map.gz emd_36800_half_map_1.map.gz emd_36800_half_map_1.map.gz emd_36800_half_map_2.map.gz emd_36800_half_map_2.map.gz | 204 MB 203.8 MB 199.9 MB 199.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36800 http://ftp.pdbj.org/pub/emdb/structures/EMD-36800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36800 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36800 | HTTPS FTP |
-Validation report
| Summary document |  emd_36800_validation.pdf.gz emd_36800_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36800_full_validation.pdf.gz emd_36800_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_36800_validation.xml.gz emd_36800_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_36800_validation.cif.gz emd_36800_validation.cif.gz | 28.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36800 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36800 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36800 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36800 | HTTPS FTP |
-Related structure data
| Related structure data |  8k1sMC  8k16C  8k1kC  8k1tC  8k1uC  8xmhC  8xmiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36800.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36800.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map for KtrAB | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_36800_msk_1.map emd_36800_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
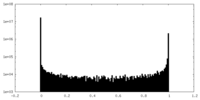
| Density Histograms |
-Additional map: focused refinement map for KtrA
| File | emd_36800_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement map for KtrA | ||||||||||||
| Projections & Slices |
| ||||||||||||
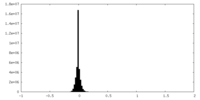
| Density Histograms |
-Additional map: focused refinement map for KtrB
| File | emd_36800_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement map for KtrB | ||||||||||||
| Projections & Slices |
| ||||||||||||
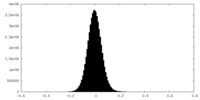
| Density Histograms |
-Half map: half map A for KtrAB
| File | emd_36800_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A for KtrAB | ||||||||||||
| Projections & Slices |
| ||||||||||||
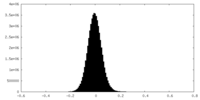
| Density Histograms |
-Half map: half map B for KtrAB
| File | emd_36800_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B for KtrAB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Bacillus subtilis KtrAB potassium transporter
| Entire | Name: Bacillus subtilis KtrAB potassium transporter |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus subtilis KtrAB potassium transporter
| Supramolecule | Name: Bacillus subtilis KtrAB potassium transporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: KtrAB complex(chain A-L). KtrAB complex in solution was composed of one KtrA octamer (chain A-H) and two KtrB dimer (chain I-J and chain K-L). The molecule weight of KtrAB complex (chain A-L) is 0.393 MDa |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 199 KDa |
-Supramolecule #2: KtrA octamer
| Supramolecule | Name: KtrA octamer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: KtrA ocatmer (chain A-H) The molecule weight of KtrA octamer (chain A-H) is 0.199 MDa. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: KtrB dimer
| Supramolecule | Name: KtrB dimer / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 Details: Two KtrB dimers (chain I-J and chain K-L) The molecule weight of each KtrB dimer is 0.097 MDa |
|---|
-Macromolecule #1: Ktr system potassium uptake protein A
| Macromolecule | Name: Ktr system potassium uptake protein A / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 24.91676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGRIKNKQFA VIGLGRFGGS ICKELHRMGH EVLAVDINEE KVNAYASYAT HAVIANATEE NELLSLGIRN FEYVIVAIGA NIQASTLTT LLLKELDIPN IWVKAQNYYH HKVLEKIGAD RIIHPEKDMG VKIAQSLSDE NVLNYIDLSD EYSIVELLAT R KLDSKSII ...String: MGRIKNKQFA VIGLGRFGGS ICKELHRMGH EVLAVDINEE KVNAYASYAT HAVIANATEE NELLSLGIRN FEYVIVAIGA NIQASTLTT LLLKELDIPN IWVKAQNYYH HKVLEKIGAD RIIHPEKDMG VKIAQSLSDE NVLNYIDLSD EYSIVELLAT R KLDSKSII DLNVRAKYGC TILAIKHHGD ICLSPAPEDI IREQDCLVIM GHKKDIKRFE NEGM UniProtKB: Ktr system potassium uptake protein A |
-Macromolecule #2: Ktr system potassium uptake protein B
| Macromolecule | Name: Ktr system potassium uptake protein B / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.471539 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTLQKDKVIK WVRFTPPQVL AIGFFLTIII GAVLLMLPIS TTKPLSWIDA LFTAASATTV TGLAVVDTGT QFTVFGQTVI MGLIQIGGL GFMTFAVLIV MILGKKIGLK ERMLVQEALN QPTIGGVIGL VKVLFLFSIS IELIAALILS IRLVPQYGWS S GLFASLFH ...String: MTLQKDKVIK WVRFTPPQVL AIGFFLTIII GAVLLMLPIS TTKPLSWIDA LFTAASATTV TGLAVVDTGT QFTVFGQTVI MGLIQIGGL GFMTFAVLIV MILGKKIGLK ERMLVQEALN QPTIGGVIGL VKVLFLFSIS IELIAALILS IRLVPQYGWS S GLFASLFH AISAFNNAGF SLWPDNLMSY VGDPTVNLVI TFLFITGGIG FTVLFDVMKN RRFKTFSLHT KLMLTGTLML NA IAMLTVF ILEYSNPGTL GHLHIVDKLW ASYFQAVTPR TAGFNSLDFG SMREGTIVFT LLLMFIGAGS ASTASGIKLT TFI VILTSV IAYLRGKKET VIFRRSIKYP IIIKALAVSV TSLFIVFLGI FALTITEQAP FLQIVFETFS AFGTVGLTMG LTPE LTTAG KCIIIVIMFI GRIGPLTFVF SFAKTEQSNI RYPDGEVFTG UniProtKB: Ktr system potassium uptake protein B |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 8 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #4: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.24 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20 mM Tris-HCl pH 8.0, 70 mM NaCl, 30 mM KCl, 0.75 mM 6-cyclohexyl-1-hexyl-beta-D-maltoside, 0.1 mM ADP |
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 15 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 6919 / Average exposure time: 2.5 sec. / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)