[English] 日本語
 Yorodumi
Yorodumi- EMDB-36765: Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, in its dual-ternary state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / hydroxylase / collagen / ER protein / CYTOSOLIC PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprocollagen-proline 3-dioxygenase / procollagen-proline 3-dioxygenase activity / protein hydroxylation / negative regulation of post-translational protein modification / endoplasmic reticulum chaperone complex / positive regulation by host of viral genome replication / Collagen biosynthesis and modifying enzymes / collagen metabolic process / positive regulation by host of viral process / positive regulation of multicellular organism growth ...procollagen-proline 3-dioxygenase / procollagen-proline 3-dioxygenase activity / protein hydroxylation / negative regulation of post-translational protein modification / endoplasmic reticulum chaperone complex / positive regulation by host of viral genome replication / Collagen biosynthesis and modifying enzymes / collagen metabolic process / positive regulation by host of viral process / positive regulation of multicellular organism growth / L-ascorbic acid binding / collagen fibril organization / RNA polymerase binding / cyclosporin A binding / regulation of protein secretion / protein peptidyl-prolyl isomerization / smooth endoplasmic reticulum / chaperone-mediated protein folding / neutrophil chemotaxis / peptidylprolyl isomerase / peptidyl-prolyl cis-trans isomerase activity / bone development / positive regulation of neuron projection development / SARS-CoV-1 activates/modulates innate immune responses / unfolded protein binding / melanosome / protein folding / spermatogenesis / protein stabilization / iron ion binding / negative regulation of cell population proliferation / endoplasmic reticulum lumen / intracellular membrane-bounded organelle / focal adhesion / perinuclear region of cytoplasm / endoplasmic reticulum / protein-containing complex / RNA binding / extracellular space / extracellular exosome / nucleoplasm / membrane / nucleus / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
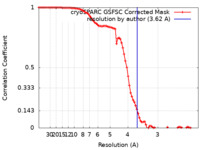
| Method | single particle reconstruction / cryo EM / Resolution: 3.62 Å | |||||||||
 Authors Authors | Li W / Peng J / Yao D / Rao B / Xia Y / Wang Q / Li S / Cao M / Shen Y / Ma P ...Li W / Peng J / Yao D / Rao B / Xia Y / Wang Q / Li S / Cao M / Shen Y / Ma P / Liao R / Qin A / Zhao J / Cao Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex. Authors: Wenguo Li / Junjiang Peng / Deqiang Yao / Bing Rao / Ying Xia / Qian Wang / Shaobai Li / Mi Cao / Yafeng Shen / Peixiang Ma / Rijing Liao / An Qin / Jie Zhao / Yu Cao /  Abstract: Collagen posttranslational processing is crucial for its proper assembly and function. Disruption of collagen processing leads to tissue development and structure disorders like osteogenesis ...Collagen posttranslational processing is crucial for its proper assembly and function. Disruption of collagen processing leads to tissue development and structure disorders like osteogenesis imperfecta (OI). OI-related collagen processing machinery includes prolyl 3-hydroxylase 1 (P3H1), peptidyl-prolyl cis-trans isomerase B (PPIB), and cartilage-associated protein (CRTAP), with their structural organization and mechanism unclear. We determine cryo-EM structures of the P3H1/CRTAP/PPIB complex. The active sites of P3H1 and PPIB form a face-to-face bifunctional reaction center, indicating a coupled modification mechanism. The structure of the P3H1/CRTAP/PPIB/collagen peptide complex reveals multiple binding sites, suggesting a substrate interacting zone. Unexpectedly, a dual-ternary complex is observed, and the balance between ternary and dual-ternary states can be altered by mutations in the P3H1/PPIB active site and the addition of PPIB inhibitors. These findings provide insights into the structural basis of collagen processing by P3H1/CRTAP/PPIB and the molecular pathology of collagen-related disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36765.map.gz emd_36765.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36765-v30.xml emd-36765-v30.xml emd-36765.xml emd-36765.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36765_fsc.xml emd_36765_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_36765.png emd_36765.png | 78.8 KB | ||
| Filedesc metadata |  emd-36765.cif.gz emd-36765.cif.gz | 6.5 KB | ||
| Others |  emd_36765_half_map_1.map.gz emd_36765_half_map_1.map.gz emd_36765_half_map_2.map.gz emd_36765_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36765 http://ftp.pdbj.org/pub/emdb/structures/EMD-36765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36765 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36765 | HTTPS FTP |
-Validation report
| Summary document |  emd_36765_validation.pdf.gz emd_36765_validation.pdf.gz | 826.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36765_full_validation.pdf.gz emd_36765_full_validation.pdf.gz | 825.9 KB | Display | |
| Data in XML |  emd_36765_validation.xml.gz emd_36765_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_36765_validation.cif.gz emd_36765_validation.cif.gz | 21.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36765 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36765 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36765 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36765 | HTTPS FTP |
-Related structure data
| Related structure data |  8k0iMC  8k0eC  8k0fC  8k0mC  8k17C  8kc9C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36765.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36765.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36765_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36765_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : A protein modification hexamer complex
| Entire | Name: A protein modification hexamer complex |
|---|---|
| Components |
|
-Supramolecule #1: A protein modification hexamer complex
| Supramolecule | Name: A protein modification hexamer complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Prolyl 3-hydroxylase 1
| Macromolecule | Name: Prolyl 3-hydroxylase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: procollagen-proline 3-dioxygenase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 83.485727 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAVRALKLLT TLLAVVAAAS QAEVESEAGW GMVTPDLLFA EGTAAYARGD WPGVVLSMER ALRSRAALRA LRLRCRTQCA ADFPWELDP DWSPSPAQAS GAAALRDLSF FGGLLRRAAC LRRCLGPPAA HSLSEEMELE FRKRSPYNYL QVAYFKINKL E KAVAAAHT ...String: MAVRALKLLT TLLAVVAAAS QAEVESEAGW GMVTPDLLFA EGTAAYARGD WPGVVLSMER ALRSRAALRA LRLRCRTQCA ADFPWELDP DWSPSPAQAS GAAALRDLSF FGGLLRRAAC LRRCLGPPAA HSLSEEMELE FRKRSPYNYL QVAYFKINKL E KAVAAAHT FFVGNPEHME MQQNLDYYQT MSGVKEADFK DLETQPHMQE FRLGVRLYSE EQPQEAVPHL EAALQEYFVA YE ECRALCE GPYDYDGYNY LEYNADLFQA ITDHYIQVLN CKQNCVTELA SHPSREKPFE DFLPSHYNYL QFAYYNIGNY TQA VECAKT YLLFFPNDEV MNQNLAYYAA MLGEEHTRSI GPRESAKEYR QRSLLEKELL FFAYDVFGIP FVDPDSWTPE EVIP KRLQE KQKSERETAV RISQEIGNLM KEIETLVEEK TKESLDVSRL TREGGPLLYE GISLTMNSKL LNGSQRVVMD GVISD HECQ ELQRLTNVAA TSGDGYRGQT SPHTPNEKFY GVTVFKALKL GQEGKVPLQS AHLYYNVTEK VRRIMESYFR LDTPLY FSY SHLVCRTAIE EVQAERKDDS HPVHVDNCIL NAETLVCVKE PPAYTFRDYS AILYLNGDFD GGNFYFTELD AKTVTAE VQ PQCGRAVGFS SGTENPHGVK AVTRGQRCAI ALWFTLDPRH SERDRVQADD LVKMLFSPEE MDLSQEQPLD AQQGPPEP A QESLSGSESK PKDEL UniProtKB: Prolyl 3-hydroxylase 1 |
-Macromolecule #2: Cartilage-associated protein
| Macromolecule | Name: Cartilage-associated protein / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50.749184 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEPGRRGAAA LLALLCVACA LRAGRAQYER YSFRSFPRDE LMPLESAYRH ALDKYSGEHW AESVGYLEIS LRLHRLLRDS EAFCHRNCS AAPQPEPAAG LASYPELRLF GGLLRRAHCL KRCKQGLPAF RQSQPSREVL ADFQRREPYK FLQFAYFKAN N LPKAIAAA ...String: MEPGRRGAAA LLALLCVACA LRAGRAQYER YSFRSFPRDE LMPLESAYRH ALDKYSGEHW AESVGYLEIS LRLHRLLRDS EAFCHRNCS AAPQPEPAAG LASYPELRLF GGLLRRAHCL KRCKQGLPAF RQSQPSREVL ADFQRREPYK FLQFAYFKAN N LPKAIAAA HTFLLKHPDD EMMKRNMAYY KSLPGAEDYI KDLETKSYES LFIRAVRAYN GENWRTSITD MELALPDFFK AF YECLAAC EGSREIKDFK DFYLSIADHY VEVLECKIQC EENLTPVIGG YPVEKFVATM YHYLQFAYYK LNDLKNAAPC AVS YLLFDQ NDKVMQQNLV YYQYHRDTWG LSDEHFQPRP EAVQFFNVTT LQKELYDFAK ENIMDDDEGE VVEYVDDLLE LEET SAAAL EVLFQGPSAW SHPQFEKGGG SGGGSGGSAW SHPQFEK UniProtKB: Cartilage-associated protein |
-Macromolecule #3: Peptidyl-prolyl cis-trans isomerase B
| Macromolecule | Name: Peptidyl-prolyl cis-trans isomerase B / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO / EC number: peptidylprolyl isomerase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 28.669605 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLRLSERNMK VLLAAALIAG SVFFLLLPGP SAADEKKKGP KVTVKVYFDL RIGDEDVGRV IFGLFGKTVP KTVDNFVALA TGEKGFGYK NSKFHRVIKD FMIQGGDFTR GDGTGGKSIY GERFPDENFK LKHYGPGWVS MANAGKDTNG SQFFITTVKT A WLDGKHVV ...String: MLRLSERNMK VLLAAALIAG SVFFLLLPGP SAADEKKKGP KVTVKVYFDL RIGDEDVGRV IFGLFGKTVP KTVDNFVALA TGEKGFGYK NSKFHRVIKD FMIQGGDFTR GDGTGGKSIY GERFPDENFK LKHYGPGWVS MANAGKDTNG SQFFITTVKT A WLDGKHVV FGKVLEGMEV VRKVESTKTD SRDKPLKDVI IADCGKIEVE KPFAIAKEAA AENLYFQGDY KDHDGDYKDH DI DYKDDDD KHHHHHHHH UniProtKB: Peptidyl-prolyl cis-trans isomerase B |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 6 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)