+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Langya Henipavirus attachment protein | |||||||||
 Map data Map data | The LayV-G protein map file. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Langya Henipavirus attachment protein / G protein / homotetramer / Viral surface glycoprotein. / VIRAL PROTEIN | |||||||||
| Biological species |  Henipavirus Henipavirus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Yan RH / Guo YY | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: A special homotetrameric architecture of langya virus glycoprotein Authors: Yan RH / Guo YY | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36741.map.gz emd_36741.map.gz | 108.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36741-v30.xml emd-36741-v30.xml emd-36741.xml emd-36741.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36741.png emd_36741.png | 94.9 KB | ||
| Filedesc metadata |  emd-36741.cif.gz emd-36741.cif.gz | 5.7 KB | ||
| Others |  emd_36741_additional_1.map.gz emd_36741_additional_1.map.gz emd_36741_half_map_1.map.gz emd_36741_half_map_1.map.gz emd_36741_half_map_2.map.gz emd_36741_half_map_2.map.gz | 185.3 MB 200.6 MB 200.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36741 http://ftp.pdbj.org/pub/emdb/structures/EMD-36741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36741 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36741 | HTTPS FTP |
-Validation report
| Summary document |  emd_36741_validation.pdf.gz emd_36741_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36741_full_validation.pdf.gz emd_36741_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_36741_validation.xml.gz emd_36741_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_36741_validation.cif.gz emd_36741_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36741 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36741 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36741 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36741.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36741.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The LayV-G protein map file. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.095 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: focused transmembrane map
| File | emd_36741_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused transmembrane map | ||||||||||||
| Projections & Slices |
| ||||||||||||
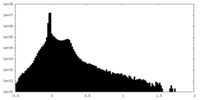
| Density Histograms |
-Half map: #2
| File | emd_36741_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
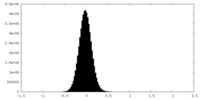
| Density Histograms |
-Half map: #1
| File | emd_36741_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homotetramer of Langya virus attachment protein G
| Entire | Name: Homotetramer of Langya virus attachment protein G |
|---|---|
| Components |
|
-Supramolecule #1: Homotetramer of Langya virus attachment protein G
| Supramolecule | Name: Homotetramer of Langya virus attachment protein G / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Henipavirus Henipavirus |
| Molecular weight | Theoretical: 270 kDa/nm |
-Macromolecule #1: langya virus attachment protein G
| Macromolecule | Name: langya virus attachment protein G / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Henipavirus Henipavirus |
| Molecular weight | Theoretical: 69.064383 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATNKRDVIK TTESTREDKV KKFYGVETAE KVADSISSNK VFILMNTLLI LTGAIITITL NVTNLTTIKN QQAMLKIIQD EVNSKLEMF VSLDQLVKGE IKPKVSLINT AVSVSIPGQI SNLQTKFLQK YVYLEESITK QCTCNPLSGI FPTTKPPPPP T DKPDDDTT ...String: MATNKRDVIK TTESTREDKV KKFYGVETAE KVADSISSNK VFILMNTLLI LTGAIITITL NVTNLTTIKN QQAMLKIIQD EVNSKLEMF VSLDQLVKGE IKPKVSLINT AVSVSIPGQI SNLQTKFLQK YVYLEESITK QCTCNPLSGI FPTTKPPPPP T DKPDDDTT DDDKVDTTIK PVEYYKPDGC NKTNDHFTMQ PGVNFYTVPN LGPSSSSADE CYTNPSFSIG SSIYMFSQEI RK TDCTTGE ILSIQIVLGR IVDKGQQGPQ ASPLLVWSVP NPKIINSCAV AAGDETGWVL CSVTLTAASG EPIPHMFDGF WLY KFEPDT EVVAYRITGF AYLLDKVYDS VFIGKGGGIQ RGNDLYFQMF GLSRNRQSIK ALCEHGSCLG TGGGGYQVLC DRAV MSFGS EESLISNAYL KVNDVASGKP TIISQTFPPS DSYKGSNGRI YTIGERYGIY LAPSSWNRYL RFGLTPDISV RSTTW LKEK DPIMKVLTTC TNTDKDMCPE ICNTRGYQDI FPLSEDSSFY TYIGITPSNE GTKSFVAVKD DAGHVASITI LPNYYS ITS ATISCFMYKE EIWCIAVTEG RKQKENPQRI YAHSYRVQKM CFNIKPASVV TSLPSNVTIR S |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 6 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source: OTHER |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.76 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 8000 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X