+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Symbiodinium photosystem I | |||||||||
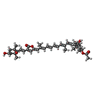
 Map data Map data | Cryo-EM structure of Symbiodinium photosystem I | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Symbiodinium / Photosystem I / PHOTOSYNTHESIS | |||||||||
| Biological species |  Symbiodinium sp. (eukaryote) Symbiodinium sp. (eukaryote) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Zhao LS / Wang N / Li K / Zhang YZ / Liu LN | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Architecture of symbiotic dinoflagellate photosystem I-light-harvesting supercomplex in Symbiodinium. Authors: Long-Sheng Zhao / Ning Wang / Kang Li / Chun-Yang Li / Jian-Ping Guo / Fei-Yu He / Gui-Ming Liu / Xiu-Lan Chen / Jun Gao / Lu-Ning Liu / Yu-Zhong Zhang /   Abstract: Symbiodinium are the photosynthetic endosymbionts for corals and play a vital role in supplying their coral hosts with photosynthetic products, forming the nutritional foundation for high-yield coral ...Symbiodinium are the photosynthetic endosymbionts for corals and play a vital role in supplying their coral hosts with photosynthetic products, forming the nutritional foundation for high-yield coral reef ecosystems. Here, we determine the cryo-electron microscopy structure of Symbiodinium photosystem I (PSI) supercomplex with a PSI core composed of 13 subunits including 2 previously unidentified subunits, PsaT and PsaU, as well as 13 peridinin-Chl a/c-binding light-harvesting antenna proteins (AcpPCIs). The PSI-AcpPCI supercomplex exhibits distinctive structural features compared to their red lineage counterparts, including extended termini of PsaD/E/I/J/L/M/R and AcpPCI-1/3/5/7/8/11 subunits, conformational changes in the surface loops of PsaA and PsaB subunits, facilitating the association between the PSI core and peripheral antennae. Structural analysis and computational calculation of excitation energy transfer rates unravel specific pigment networks in Symbiodinium PSI-AcpPCI for efficient excitation energy transfer. Overall, this study provides a structural basis for deciphering the mechanisms governing light harvesting and energy transfer in Symbiodinium PSI-AcpPCI supercomplexes adapted to their symbiotic ecosystem, as well as insights into the evolutionary diversity of PSI-LHCI among various photosynthetic organisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36366.map.gz emd_36366.map.gz | 112.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36366-v30.xml emd-36366-v30.xml emd-36366.xml emd-36366.xml | 42.7 KB 42.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36366.png emd_36366.png | 192.9 KB | ||
| Filedesc metadata |  emd-36366.cif.gz emd-36366.cif.gz | 11 KB | ||
| Others |  emd_36366_half_map_1.map.gz emd_36366_half_map_1.map.gz emd_36366_half_map_2.map.gz emd_36366_half_map_2.map.gz | 115.9 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36366 http://ftp.pdbj.org/pub/emdb/structures/EMD-36366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36366 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36366 | HTTPS FTP |
-Validation report
| Summary document |  emd_36366_validation.pdf.gz emd_36366_validation.pdf.gz | 981.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36366_full_validation.pdf.gz emd_36366_full_validation.pdf.gz | 981.2 KB | Display | |
| Data in XML |  emd_36366_validation.xml.gz emd_36366_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_36366_validation.cif.gz emd_36366_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36366 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36366 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36366 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36366 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36366.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36366.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Symbiodinium photosystem I | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
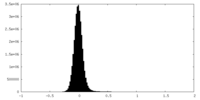
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Cryo-EM structure of Symbiodinium photosystem I
| File | emd_36366_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Symbiodinium photosystem I | ||||||||||||
| Projections & Slices |
| ||||||||||||
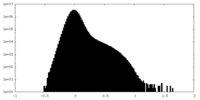
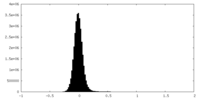
| Density Histograms |
-Half map: Cryo-EM structure of Symbiodinium photosystem I
| File | emd_36366_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of Symbiodinium photosystem I | ||||||||||||
| Projections & Slices |
| ||||||||||||
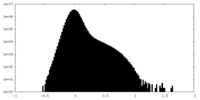
| Density Histograms |
- Sample components
Sample components
+Entire : Photosystem I of Symbiodinium
+Supramolecule #1: Photosystem I of Symbiodinium
+Macromolecule #1: PCPI-7
+Macromolecule #2: PCPI-1
+Macromolecule #3: PCP-11
+Macromolecule #4: PCPI-6
+Macromolecule #5: PCPI-5
+Macromolecule #6: PCPI-8
+Macromolecule #7: PCPI-4
+Macromolecule #8: PCPI-10
+Macromolecule #9: PCPI-3
+Macromolecule #10: PCPI-9
+Macromolecule #11: PCPI-13
+Macromolecule #12: PCPI-12
+Macromolecule #13: PCPI-2
+Macromolecule #14: PsaA
+Macromolecule #15: PsaB
+Macromolecule #16: PsaC
+Macromolecule #17: PsaD
+Macromolecule #18: PsaE
+Macromolecule #19: PsaF
+Macromolecule #20: PsaI
+Macromolecule #21: PsaJ
+Macromolecule #22: PsaL
+Macromolecule #23: PsaM
+Macromolecule #24: PsaR
+Macromolecule #25: PsaT
+Macromolecule #26: PsaU
+Macromolecule #27: CHLOROPHYLL A
+Macromolecule #28: Chlorophyll c2
+Macromolecule #29: [(1~{S},5~{R})-3,3,5-trimethyl-5-oxidanyl-4-[(3~{E},5~{E},7~{E},9...
+Macromolecule #30: (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,be...
+Macromolecule #31: 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL
+Macromolecule #32: PERIDININ
+Macromolecule #33: 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE
+Macromolecule #34: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
+Macromolecule #35: DIGALACTOSYL DIACYL GLYCEROL (DGDG)
+Macromolecule #36: DODECYL-ALPHA-D-MALTOSIDE
+Macromolecule #37: PHYLLOQUINONE
+Macromolecule #38: BETA-CAROTENE
+Macromolecule #39: IRON/SULFUR CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 161863 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)