[English] 日本語
 Yorodumi
Yorodumi- EMDB-36116: Cryo-EM structure of the African swine fever virus topoisomerase ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide (EDI-1) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Topoisomerase / ASFV / inhibitor / VIRAL PROTEIN / ISOMERASE-DNA complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationsister chromatid segregation / DNA topoisomerase type II (double strand cut, ATP-hydrolyzing) activity / DNA topoisomerase (ATP-hydrolysing) / DNA topological change / DNA binding / ATP binding / metal ion binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |   African swine fever virus / African swine fever virus /  Homo sapiens (human) Homo sapiens (human) | |||||||||
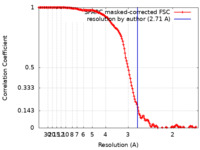
| Method | single particle reconstruction / cryo EM / Resolution: 2.71 Å | |||||||||
 Authors Authors | Chang C-W / Tsai M-D | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Chem / Year: 2024 Journal: Commun Chem / Year: 2024Title: A unified view on enzyme catalysis by cryo-EM study of a DNA topoisomerase. Authors: Chiung-Wen Mary Chang / Shun-Chang Wang / Chun-Hsiung Wang / Allan H Pang / Cheng-Han Yang / Yao-Kai Chang / Wen-Jin Wu / Ming-Daw Tsai /   Abstract: The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the ...The theories for substrate recognition in enzyme catalysis have evolved from lock-key to induced fit, then conformational selection, and conformational selection followed by induced fit. However, the prevalence and consensus of these theories require further examination. Here we use cryogenic electron microscopy and African swine fever virus type 2 topoisomerase (AsfvTop2) to demonstrate substrate binding theories in a joint and ordered manner: catalytic selection by the enzyme, conformational selection by the substrates, then induced fit. The apo-AsfvTop2 pre-exists in six conformers that comply with the two-gate mechanism directing DNA passage and release in the Top2 catalytic cycle. The structures of AsfvTop2-DNA-inhibitor complexes show that substantial induced-fit changes occur locally from the closed apo-conformer that however is too far-fetched for the open apo-conformer. Furthermore, the ATPase domain of AsfvTop2 in the MgAMP-PNP-bound crystal structures coexist in reduced and oxidized forms involving a disulfide bond, which can regulate the AsfvTop2 function. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36116.map.gz emd_36116.map.gz | 153.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36116-v30.xml emd-36116-v30.xml emd-36116.xml emd-36116.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36116_fsc.xml emd_36116_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_36116.png emd_36116.png | 90.3 KB | ||
| Filedesc metadata |  emd-36116.cif.gz emd-36116.cif.gz | 6.8 KB | ||
| Others |  emd_36116_half_map_1.map.gz emd_36116_half_map_1.map.gz emd_36116_half_map_2.map.gz emd_36116_half_map_2.map.gz | 285 MB 285 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36116 http://ftp.pdbj.org/pub/emdb/structures/EMD-36116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36116 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36116 | HTTPS FTP |
-Validation report
| Summary document |  emd_36116_validation.pdf.gz emd_36116_validation.pdf.gz | 706.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36116_full_validation.pdf.gz emd_36116_full_validation.pdf.gz | 706 KB | Display | |
| Data in XML |  emd_36116_validation.xml.gz emd_36116_validation.xml.gz | 23.6 KB | Display | |
| Data in CIF |  emd_36116_validation.cif.gz emd_36116_validation.cif.gz | 30.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36116 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36116 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36116 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36116 | HTTPS FTP |
-Related structure data
| Related structure data |  8j9vMC  8j87C  8j88C  8j89C  8j8aC  8j8bC  8j8cC  8j9wC  8j9xC  8ja1C  8ja2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36116.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36116.map.gz / Format: CCP4 / Size: 307.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36116_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36116_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : African swine fever virus topoisomerase 2 complexed with Cut02aDN...
| Entire | Name: African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide |
|---|---|
| Components |
|
-Supramolecule #1: African swine fever virus topoisomerase 2 complexed with Cut02aDN...
| Supramolecule | Name: African swine fever virus topoisomerase 2 complexed with Cut02aDNA and etoposide type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:   African swine fever virus African swine fever virus |
-Macromolecule #1: DNA topoisomerase 2
| Macromolecule | Name: DNA topoisomerase 2 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   African swine fever virus African swine fever virus |
| Molecular weight | Theoretical: 136.422578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEAFEISDFK EHAKKKSMWA GALNKVTISG LMGVFTEDED LMALPIHRDH CPALLKIFDE LIVNATDHER ACHSKTKKVT YIKISFDKG VFSCENDGPG IPIAKHEQAS LIAKRDVYVP EVASCFFLAG TNINKAKDCI KGGTNGVGLK LAMVHSQWAI L TTADGAQK ...String: MEAFEISDFK EHAKKKSMWA GALNKVTISG LMGVFTEDED LMALPIHRDH CPALLKIFDE LIVNATDHER ACHSKTKKVT YIKISFDKG VFSCENDGPG IPIAKHEQAS LIAKRDVYVP EVASCFFLAG TNINKAKDCI KGGTNGVGLK LAMVHSQWAI L TTADGAQK YVQQINQRLD IIEPPTITPS REMFTRIELM PVYQELGYAE PLSETEQADL SAWIYLRACQ CAAYVGKGTT IY YNDKPCR TGSVMALAKM YTLLSAPNST IHTATIKADA KPYSLHPLQV AAVVSPKFKK FEHVSIINGV NCVKGEHVTF LKK TINEMV IKKFQQTIKD KNRKTTLRDS CSNIFVVIVG SIPGIEWTGQ RKDELSIAEN VFKTHYSIPS SFLTSMTRSI VDIL LQSIS KKDNHKQVDV DKYTRARNAG GKRAQDCMLL AAEGDSALSL LRTGLTLGKS NPSGPSFDFC GMISLGGVIM NACKK VTNI TTDSGETIMV RNEQLTNNKV LQGIVQVLGL DFNCHYKTQE ERAKLRYGCI VACVDQDLDG CGKILGLLLA YFHLFW PQL IIHGFVKRLL TPLIRVYEKG KTMPVEFYYE QEFDAWAKKQ TSLVNHTVKY YKGLAAHDTH EVKSMFKHFD NMVYTFT LD DSAKELFHIY FGGESELRKR ELCTGVVPLT ETQTQSIHSV RRIPCSLHLQ VDTKAYKLDA IERQIPNFLD GMTRARRK I LAGGVKCFAS NNRERKVFQF GGYVADHMFY HHGDMSLNTS IIKAAQYYPG SSHLYPVFIG IGSFGSRHLG GKDAGSPRY ISVQLASEFI KTMFPAEDSW LLPYVFEDGQ RAEPEYYVPV LPLAIMEYGA NPSEGWKYTT WARQLEDILA LVRAYVDKDN PKHELLHYA IKHKITILPL RPSNYNFKGH LKRFGQYYYS YGTYDISEQR NIITITELPL RVPTVAYIES IKKSSNRMTF I EEIIDYSS SETIEILVKL KPNSLNRIVE EFKETEEQDS IENFLRLRNC LHSHLNFVKP KGGIIEFNSY YEILYAWLPY RR ELYQKRL MREHAVLKLR IIMETAIVRY INESAELNLS HYEDEKEASR ILSEHGFPPL NHTLIISPEF ASIEELNQKA LQG CYTYIL SLQARELLIA AKTRRVEKIK KMQARLDKVE QLLQESPFPG ASVWLEEIDA VEKAIIKGRN TQWKFHHHHH H UniProtKB: DNA topoisomerase 2 |
-Macromolecule #2: DNA (5'-D(*GP*AP*GP*GP*TP*AP*TP*GP*TP*AP*GP*GP*C)-3')
| Macromolecule | Name: DNA (5'-D(*GP*AP*GP*GP*TP*AP*TP*GP*TP*AP*GP*GP*C)-3') / type: dna / ID: 2 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.071657 KDa |
| Sequence | String: (DG)(DA)(DG)(DG)(DT)(DA)(DT)(DG)(DT)(DA) (DG)(DG)(DC) |
-Macromolecule #3: DNA (5'-D(*GP*GP*CP*CP*GP*CP*CP*TP*AP*CP*AP*TP*AP*CP*CP*TP*C)-3')
| Macromolecule | Name: DNA (5'-D(*GP*GP*CP*CP*GP*CP*CP*TP*AP*CP*AP*TP*AP*CP*CP*TP*C)-3') type: dna / ID: 3 / Number of copies: 2 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5.108312 KDa |
| Sequence | String: (DG)(DG)(DC)(DC)(DG)(DC)(DC)(DT)(DA)(DC) (DA)(DT)(DA)(DC)(DC)(DT)(DC) |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #5: (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,...
| Macromolecule | Name: (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside type: ligand / ID: 5 / Number of copies: 2 / Formula: EVP |
|---|---|
| Molecular weight | Theoretical: 588.557 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 11786 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT | |||||||||
| Output model |  PDB-8j9v: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)