+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Membrane bound PRTase, C3 symmetry, donor bound | |||||||||
 Map data Map data | EM map of a membrane-bound PRTase, C3 symmetry donor-bound | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | phosphoribose transferase complex / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationdecaprenyl-phosphate phosphoribosyltransferase / arabinosyltransferase activity / glycolipid biosynthetic process / capsule polysaccharide biosynthetic process / transferase activity, transferring alkyl or aryl (other than methyl) groups / cell wall organization / magnesium ion binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Wu FY / Gao S / Zhang L / Rao ZH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Microbiol / Year: 2024 Journal: Nat Microbiol / Year: 2024Title: Structural analysis of phosphoribosyltransferase-mediated cell wall precursor synthesis in Mycobacterium tuberculosis. Authors: Shan Gao / Fangyu Wu / Sudagar S Gurcha / Sarah M Batt / Gurdyal S Besra / Zihe Rao / Lu Zhang /   Abstract: In Mycobacterium tuberculosis, Rv3806c is a membrane-bound phosphoribosyltransferase (PRTase) involved in cell wall precursor production. It catalyses pentosyl phosphate transfer from phosphoribosyl ...In Mycobacterium tuberculosis, Rv3806c is a membrane-bound phosphoribosyltransferase (PRTase) involved in cell wall precursor production. It catalyses pentosyl phosphate transfer from phosphoribosyl pyrophosphate to decaprenyl phosphate, to generate 5-phospho-β-ribosyl-1-phosphoryldecaprenol. Despite Rv3806c being an attractive drug target, structural and molecular mechanistic insight into this PRTase is lacking. Here we report cryogenic electron microscopy structures for Rv3806c in the donor- and acceptor-bound states. In a lipidic environment, Rv3806c is trimeric, creating a UbiA-like fold. Each protomer forms two helical bundles, which, alongside the bound lipids, are required for PRTase activity in vitro. Mutational and functional analyses reveal that decaprenyl phosphate and phosphoribosyl pyrophosphate bind the intramembrane and extramembrane cavities of Rv3806c, respectively, in a distinct manner to that of UbiA superfamily enzymes. Our data suggest a model for Rv3806c-catalysed phosphoribose transfer through an inverting mechanism. These findings provide a structural basis for cell wall precursor biosynthesis that could have potential for anti-tuberculosis drug development. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36071.map.gz emd_36071.map.gz | 106.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36071-v30.xml emd-36071-v30.xml emd-36071.xml emd-36071.xml | 18.2 KB 18.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36071.png emd_36071.png | 38.8 KB | ||
| Filedesc metadata |  emd-36071.cif.gz emd-36071.cif.gz | 6.4 KB | ||
| Others |  emd_36071_half_map_1.map.gz emd_36071_half_map_1.map.gz emd_36071_half_map_2.map.gz emd_36071_half_map_2.map.gz | 104.8 MB 104.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36071 http://ftp.pdbj.org/pub/emdb/structures/EMD-36071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36071 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36071 | HTTPS FTP |
-Validation report
| Summary document |  emd_36071_validation.pdf.gz emd_36071_validation.pdf.gz | 715.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36071_full_validation.pdf.gz emd_36071_full_validation.pdf.gz | 714.9 KB | Display | |
| Data in XML |  emd_36071_validation.xml.gz emd_36071_validation.xml.gz | 13.8 KB | Display | |
| Data in CIF |  emd_36071_validation.cif.gz emd_36071_validation.cif.gz | 16.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36071 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36071 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36071 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36071 | HTTPS FTP |
-Related structure data
| Related structure data |  8j8jMC  8j8kC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36071.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36071.map.gz / Format: CCP4 / Size: 113.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map of a membrane-bound PRTase, C3 symmetry donor-bound | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.96 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: EM half map B of a membrane-bound PRTase, C3 symmetry donor-bound
| File | emd_36071_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map B of a membrane-bound PRTase, C3 symmetry donor-bound | ||||||||||||
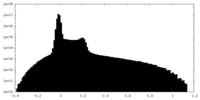
| Projections & Slices |
| ||||||||||||
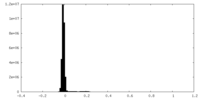
| Density Histograms |
-Half map: EM half map A of a membrane-bound PRTase, C3 symmetry donor-bound
| File | emd_36071_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM half map A of a membrane-bound PRTase, C3 symmetry donor-bound | ||||||||||||
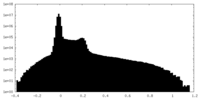
| Projections & Slices |
| ||||||||||||
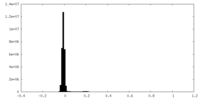
| Density Histograms |
- Sample components
Sample components
-Entire : trimeric phosphoribosyltransferase
| Entire | Name: trimeric phosphoribosyltransferase |
|---|---|
| Components |
|
-Supramolecule #1: trimeric phosphoribosyltransferase
| Supramolecule | Name: trimeric phosphoribosyltransferase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: trimeric phosphoribosyltransferase, donor bound |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
| Molecular weight | Theoretical: 97.8 KDa |
-Macromolecule #1: Decaprenyl-phosphate phosphoribosyltransferase
| Macromolecule | Name: Decaprenyl-phosphate phosphoribosyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO / EC number: decaprenyl-phosphate phosphoribosyltransferase |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) |
| Molecular weight | Theoretical: 32.682355 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MSEDVVTQPP ANLVAGVVKA IRPRQWVKNV LVLAAPLAAL GGGVRYDYVE VLSKVSMAFV VFSLAASAVY LVNDVRDVEA DREHPTKRF RPIAAGVVPE WLAYTVAVVL GVTSLAGAWM LTPNLALVMV VYLAMQLAYC FGLKHQAVVE ICVVSSAYLI R AIAGGVAT ...String: MSEDVVTQPP ANLVAGVVKA IRPRQWVKNV LVLAAPLAAL GGGVRYDYVE VLSKVSMAFV VFSLAASAVY LVNDVRDVEA DREHPTKRF RPIAAGVVPE WLAYTVAVVL GVTSLAGAWM LTPNLALVMV VYLAMQLAYC FGLKHQAVVE ICVVSSAYLI R AIAGGVAT KIPLSKWFLL IMAFGSLFMV AGKRYAELHL AERTGAAIRK SLESYTSTYL RFVWTLSATA VVLCYGLWAF ER DGYSGSW FAVSMIPFTI AILRYAVDVD GGLAGEPEDI ALRDRVLQLL ALAWIATVGA AVAFG UniProtKB: Decaprenyl-phosphate phosphoribosyltransferase |
-Macromolecule #2: 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose
| Macromolecule | Name: 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose / type: ligand / ID: 2 / Number of copies: 3 / Formula: PRP |
|---|---|
| Molecular weight | Theoretical: 390.07 Da |
| Chemical component information |  ChemComp-PRP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]o...
| Macromolecule | Name: (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate type: ligand / ID: 4 / Number of copies: 3 / Formula: PGW |
|---|---|
| Molecular weight | Theoretical: 749.007 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.0 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 40 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % | |||||||||
| Details | This sample was mono disperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: TFS Selectris |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Number real images: 9908 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z
Z Y
Y X
X