+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Composite map of the Rad51-ADP filament | |||||||||
 Map data Map data | This is a composite map with a high-resolution map (3.14 A) of the central part of the helix combined with the other part from a lower-resolution map (3.58A). | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Recombinase / ATPase / DNA repair protein / DNA binding / RECOMBINATION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 3.58 Å | |||||||||
 Authors Authors | Miki Y / Luo SC / Ho MC | |||||||||
| Funding support |  Taiwan, 1 items Taiwan, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A RAD51-ADP double filament structure unveils the mechanism of filament dynamics in homologous recombination. Authors: Shih-Chi Luo / Min-Chi Yeh / Yu-Hsiang Lien / Hsin-Yi Yeh / Huei-Lun Siao / I-Ping Tu / Peter Chi / Meng-Chiao Ho /  Abstract: ATP-dependent RAD51 recombinases play an essential role in eukaryotic homologous recombination by catalyzing a four-step process: 1) formation of a RAD51 single-filament assembly on ssDNA in the ...ATP-dependent RAD51 recombinases play an essential role in eukaryotic homologous recombination by catalyzing a four-step process: 1) formation of a RAD51 single-filament assembly on ssDNA in the presence of ATP, 2) complementary DNA strand-exchange, 3) ATP hydrolysis transforming the RAD51 filament into an ADP-bound disassembly-competent state, and 4) RAD51 disassembly to provide access for DNA repairing enzymes. Of these steps, filament dynamics between the ATP- and ADP-bound states, and the RAD51 disassembly mechanism, are poorly understood due to the lack of near-atomic-resolution information of the ADP-bound RAD51-DNA filament structure. We report the cryo-EM structure of ADP-bound RAD51-DNA filaments at 3.1 Å resolution, revealing a unique RAD51 double-filament that wraps around ssDNA. Structural analysis, supported by ATP-chase and time-resolved cryo-EM experiments, reveals a collapsing mechanism involving two four-protomer movements along ssDNA for mechanical transition between RAD51 single- and double-filament without RAD51 dissociation. This mechanism enables elastic change of RAD51 filament length during structural transitions between ATP- and ADP-states. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36040.map.gz emd_36040.map.gz | 194.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36040-v30.xml emd-36040-v30.xml emd-36040.xml emd-36040.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36040.png emd_36040.png | 2.5 MB | ||
| Others |  emd_36040_additional_1.map.gz emd_36040_additional_1.map.gz emd_36040_half_map_1.map.gz emd_36040_half_map_1.map.gz emd_36040_half_map_2.map.gz emd_36040_half_map_2.map.gz | 203.1 MB 199.2 MB 199.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36040 http://ftp.pdbj.org/pub/emdb/structures/EMD-36040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36040 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36040 | HTTPS FTP |
-Validation report
| Summary document |  emd_36040_validation.pdf.gz emd_36040_validation.pdf.gz | 868 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36040_full_validation.pdf.gz emd_36040_full_validation.pdf.gz | 867.6 KB | Display | |
| Data in XML |  emd_36040_validation.xml.gz emd_36040_validation.xml.gz | 15.7 KB | Display | |
| Data in CIF |  emd_36040_validation.cif.gz emd_36040_validation.cif.gz | 18.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36040 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36040 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36040 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36040.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36040.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is a composite map with a high-resolution map (3.14 A) of the central part of the helix combined with the other part from a lower-resolution map (3.58A). | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||
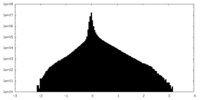
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: This is the consensus low-resolution density map (3.58...
| File | emd_36040_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is the consensus low-resolution density map (3.58 A) used for composite map generation with a high-resolution map (3.14 A) locally refined in the central part of the helix. | ||||||||||||
| Projections & Slices |
| ||||||||||||
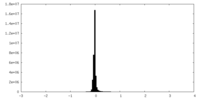
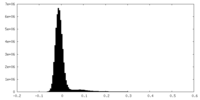
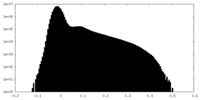
| Density Histograms |
-Half map: This is half map A of the consensus map.
| File | emd_36040_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is half map A of the consensus map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
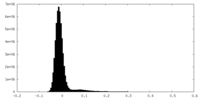
| Density Histograms |
-Half map: This is half map B of the consensus map.
| File | emd_36040_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | This is half map B of the consensus map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
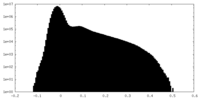
| Density Histograms |
- Sample components
Sample components
-Entire : hRAD51-ADP filament intermediate
| Entire | Name: hRAD51-ADP filament intermediate |
|---|---|
| Components |
|
-Supramolecule #1: hRAD51-ADP filament intermediate
| Supramolecule | Name: hRAD51-ADP filament intermediate / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: hRAD51 protein filament assembled in the presence of ADP and single strand DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: DNA repair protein RAD51 homolog 1
| Macromolecule | Name: DNA repair protein RAD51 homolog 1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MAMQMQLEAN ADTSVEEESF GPQPISRLEQ CGINANDVKK LEEAGFHTVE AVAYAPKKEL INIKGISEAK ADKILAEAAK LVPMGFTTA TEFHQRRSEI IQITTGSKEL DKLLQGGIET GSITEMFGEF RTGKTQICHT LAVTCQLPID RGGGEGKAMY I DTEGTFRP ...String: MAMQMQLEAN ADTSVEEESF GPQPISRLEQ CGINANDVKK LEEAGFHTVE AVAYAPKKEL INIKGISEAK ADKILAEAAK LVPMGFTTA TEFHQRRSEI IQITTGSKEL DKLLQGGIET GSITEMFGEF RTGKTQICHT LAVTCQLPID RGGGEGKAMY I DTEGTFRP ERLLAVAERY GLSGSDVLDN VAYARAFNTD HQTQLLYQAS AMMVESRYAL LIVDSATALY RTDYSGRGEL SA RQMHLAR FLRMLLRLAD EFGVAVVITN QVVAQVDGAA MFAADPKKPI GGNIIAHAST TRLYLRKGRG ETRICKIYDS PCL PEAEAM FAINADGVGD AKD |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 9.8 Å Applied symmetry - Helical parameters - Δ&Phi: 155.6 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 3.58 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 2622222 |
|---|---|
| Startup model | Type of model: INSILICO MODEL |
| Final angle assignment | Type: NOT APPLICABLE |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X