+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
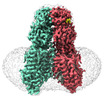
| Title | SIDT1 protein | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | transport T1 / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRNA transmembrane transporter activity / RNA transport / cholesterol binding / bioluminescence / generation of precursor metabolites and energy / double-stranded RNA binding / lysosome / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.77 Å | |||||||||
 Authors Authors | Zhang JT / Jiang DH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structural insights into double-stranded RNA recognition and transport by SID-1. Authors: Jiangtao Zhang / Chunhua Zhan / Junping Fan / Dian Wu / Ruixue Zhang / Di Wu / Xinyao Chen / Ying Lu / Ming Li / Min Lin / Jianke Gong / Daohua Jiang /  Abstract: RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA ...RNA uptake by cells is critical for RNA-mediated gene interference (RNAi) and RNA-based therapeutics. In Caenorhabditis elegans, RNAi is systemic as a result of SID-1-mediated double-stranded RNA (dsRNA) across cells. Despite the functional importance, the underlying mechanisms of dsRNA internalization by SID-1 remain elusive. Here we describe cryogenic electron microscopy structures of SID-1, SID-1-dsRNA complex and human SID-1 homologs SIDT1 and SIDT2, elucidating the structural basis of dsRNA recognition and import by SID-1. The homodimeric SID-1 homologs share conserved architecture, but only SID-1 possesses the molecular determinants within its extracellular domains for distinguishing dsRNA from single-stranded RNA and DNA. We show that the removal of the long intracellular loop between transmembrane helix 1 and 2 attenuates dsRNA uptake and systemic RNAi in vivo, suggesting a possible endocytic mechanism of SID-1-mediated dsRNA internalization. Our study provides mechanistic insights into dsRNA internalization by SID-1, which may facilitate the development of dsRNA applications based on SID-1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36008.map.gz emd_36008.map.gz | 118 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36008-v30.xml emd-36008-v30.xml emd-36008.xml emd-36008.xml | 15.6 KB 15.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36008.png emd_36008.png | 178.1 KB | ||
| Filedesc metadata |  emd-36008.cif.gz emd-36008.cif.gz | 6.1 KB | ||
| Others |  emd_36008_half_map_1.map.gz emd_36008_half_map_1.map.gz emd_36008_half_map_2.map.gz emd_36008_half_map_2.map.gz | 116 MB 115.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36008 http://ftp.pdbj.org/pub/emdb/structures/EMD-36008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36008 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36008 | HTTPS FTP |
-Validation report
| Summary document |  emd_36008_validation.pdf.gz emd_36008_validation.pdf.gz | 946.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36008_full_validation.pdf.gz emd_36008_full_validation.pdf.gz | 946.5 KB | Display | |
| Data in XML |  emd_36008_validation.xml.gz emd_36008_validation.xml.gz | 13.9 KB | Display | |
| Data in CIF |  emd_36008_validation.cif.gz emd_36008_validation.cif.gz | 16.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36008 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36008 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36008 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36008 | HTTPS FTP |
-Related structure data
| Related structure data |  8j6mMC  8hipC  8hkeC  8j6oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36008.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36008.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36008_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36008_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : T1
| Entire | Name: T1 |
|---|---|
| Components |
|
-Supramolecule #1: T1
| Supramolecule | Name: T1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Green fluorescent protein,SID1 transmembrane family member 1
| Macromolecule | Name: Green fluorescent protein,SID1 transmembrane family member 1 type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 124.044672 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MYRMQLLSCI ALSLALVTNS WSHPQFEKGG GSGGGSGGSA WSHPQFEKSK GEELFTGVVP ILVELDGDVN GHKFSVRGEG EGDATNGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKRHD FFKSAMPEGY VQERTISFKD DGTYKTRAEV K FEGDTLVN ...String: MYRMQLLSCI ALSLALVTNS WSHPQFEKGG GSGGGSGGSA WSHPQFEKSK GEELFTGVVP ILVELDGDVN GHKFSVRGEG EGDATNGKL TLKFICTTGK LPVPWPTLVT TLTYGVQCFS RYPDHMKRHD FFKSAMPEGY VQERTISFKD DGTYKTRAEV K FEGDTLVN RIELKGIDFK EDGNILGHKL EYNFNSHNVY ITADKQKNGI KANFKIRHNV EDGSVQLADH YQQNTPIGDG PV LLPDNHY LSTQSVLSKD PNEKRDHMVL LEFVTAAGIT HGMDELEVLF QGPASPGHPA KSPRQPPAPR RDPFDAARGA DFD HVYSGV VNLSTENIYS FNYTSQPDQV TAVRVYVNSS SENLNYPVLV VVRQQKEVLS WQVPLLFQGL YQRSYNYQEV SRTL CPSEA TNETGPLQQL IFVDVASMAP LGAQYKLLVT KLKHFQLRTN VAFHFTASPS QPQYFLYKFP KDVDSVIIKV VSEMA YPCS VVSVQNIMCP VYDLDHNVEF NGVYQSMTKK AAITLQKKDF PGEQFFVVFV IKPEDYACGG SFFIQEKENQ TWNLQR KKN LEVTIVPSIK ESVYVKSSLF SVFIFLSFYL GCLLVGFVHY LRFQRKSIDG SFGSNDGSGN MVASHPIAAS TPEGSNY GT IDESSSSPGR QMSSSDGGPP GQSDTDSSVE ESDFDTMPDI ESDKNIIRTK MFLYLSDLSR KDRRIVSKKY KIYFWNII T IAVFYALPVI QLVITYQTVV NVTGNQDICY YNFLCAHPLG VLSAFNNILS NLGHVLLGFL FLLIVLRRDI LHRRALEAK DIFAVEYGIP KHFGLFYAMG IALMMEGVLS ACYHVCPNYS NFQFDTSFMY MIAGLCMLKL YQTRHPDINA SAYSAYASFA VVIMVTVLG VVFGKNDVWF WVIFSAIHVL ASLALSTQIY YMGRFKIDLG IFRRAAMVFY TDCIQQCSRP LYMDRMVLLV V GNLVNWSF ALFGLIYRPR DFASYMLGIF ICNLLLYLAF YIIMKLRSSE KVLPVPLFCI VATAVMWAAA LYFFFQNLSS WE GTPAESR EKNRECILLD FFDDHDIWHF LSATALFFSF LVLLTLDDDL DVVRRDQIPV F UniProtKB: Green fluorescent protein, SID1 transmembrane family member 1 |
-Macromolecule #2: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 2 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: OLEIC ACID
| Macromolecule | Name: OLEIC ACID / type: ligand / ID: 5 / Number of copies: 2 / Formula: OLA |
|---|---|
| Molecular weight | Theoretical: 282.461 Da |
| Chemical component information |  ChemComp-OLA: |
-Macromolecule #6: water
| Macromolecule | Name: water / type: ligand / ID: 6 / Number of copies: 2 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.77 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 145765 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)