+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
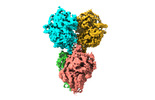
| Title | The cryo-EM structure of the TwOSC1 tetramer | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | oxidosqualene cyclase / Tripterygium wilfordii Hook. f. / TwOSC1 / friedelin / cryo-EM structure / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationbeta-amyrin synthase activity / Isomerases; Intramolecular transferases; Transferring other groups / triterpenoid biosynthetic process / lipid droplet Similarity search - Function | |||||||||
| Biological species |  Tripterygium wilfordii (plant) Tripterygium wilfordii (plant) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.75 Å | |||||||||
 Authors Authors | Ma X / Yuru T / Yunfeng L / Jiang T | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2023 Journal: Angew Chem Int Ed Engl / Year: 2023Title: Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC. Authors: Yunfeng Luo / Xiaoli Ma / Yufan Qiu / Yun Lu / Siyu Shen / Yang Li / Haiyun Gao / Kang Chen / Jiawei Zhou / Tianyuan Hu / Lichan Tu / Huan Zhao / Dan Li / Faqiang Leng / Wei Gao / Tao Jiang ...Authors: Yunfeng Luo / Xiaoli Ma / Yufan Qiu / Yun Lu / Siyu Shen / Yang Li / Haiyun Gao / Kang Chen / Jiawei Zhou / Tianyuan Hu / Lichan Tu / Huan Zhao / Dan Li / Faqiang Leng / Wei Gao / Tao Jiang / Changli Liu / Luqi Huang / Ruibo Wu / Yuru Tong /  Abstract: The oxidosqualene cyclase (OSC) catalyzed cyclization of the linear substrate (3S)-2,3-oxidosqualene to form diverse pentacyclic triterpenoid (PT) skeletons is one of the most complex reactions in ...The oxidosqualene cyclase (OSC) catalyzed cyclization of the linear substrate (3S)-2,3-oxidosqualene to form diverse pentacyclic triterpenoid (PT) skeletons is one of the most complex reactions in nature. Friedelin has a unique PT skeleton involving a fascinating nine-step cation shuttle run (CSR) cascade rearrangement reaction, in which the carbocation formed at C2 moves to the other side of the skeleton, runs back to C3 to yield a friedelin cation, which is finally deprotonated. However, as crystal structure data of plant OSCs are lacking, it remains unknown why the CSR cascade reactions occur in friedelin biosynthesis, as does the exact catalytic mechanism of the CSR. In this study, we determined the first cryogenic electron microscopy structure of a plant OSC, friedelin synthase, from Tripterygium wilfordii Hook. f (TwOSC). We also performed quantum mechanics/molecular mechanics simulations to reveal the energy profile for the CSR cascade reaction and identify key residues crucial for PT skeleton formation. Furthermore, we semirationally designed two TwOSC mutants, which significantly improved the yields of friedelin and β-amyrin, respectively. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35996.map.gz emd_35996.map.gz | 49.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35996-v30.xml emd-35996-v30.xml emd-35996.xml emd-35996.xml | 14.8 KB 14.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35996.png emd_35996.png | 52.9 KB | ||
| Filedesc metadata |  emd-35996.cif.gz emd-35996.cif.gz | 5.7 KB | ||
| Others |  emd_35996_half_map_1.map.gz emd_35996_half_map_1.map.gz emd_35996_half_map_2.map.gz emd_35996_half_map_2.map.gz | 39.7 MB 39.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35996 http://ftp.pdbj.org/pub/emdb/structures/EMD-35996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35996 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35996 | HTTPS FTP |
-Validation report
| Summary document |  emd_35996_validation.pdf.gz emd_35996_validation.pdf.gz | 949.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35996_full_validation.pdf.gz emd_35996_full_validation.pdf.gz | 949.1 KB | Display | |
| Data in XML |  emd_35996_validation.xml.gz emd_35996_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_35996_validation.cif.gz emd_35996_validation.cif.gz | 13.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35996 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35996 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35996 | HTTPS FTP |
-Related structure data
| Related structure data |  8j5zMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35996.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35996.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35996_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
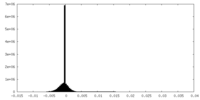
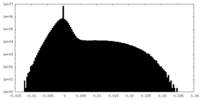
| Density Histograms |
-Half map: #1
| File | emd_35996_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
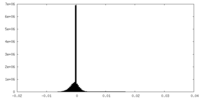
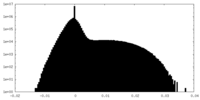
| Density Histograms |
- Sample components
Sample components
-Entire : Tetramer of TwOSC1
| Entire | Name: Tetramer of TwOSC1 |
|---|---|
| Components |
|
-Supramolecule #1: Tetramer of TwOSC1
| Supramolecule | Name: Tetramer of TwOSC1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Tripterygium wilfordii (plant) Tripterygium wilfordii (plant) |
-Macromolecule #1: Terpene cyclase/mutase family member
| Macromolecule | Name: Terpene cyclase/mutase family member / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tripterygium wilfordii (plant) Tripterygium wilfordii (plant) |
| Molecular weight | Theoretical: 88.2125 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MWKLKVAERG NAPYSEYLYT TNDFSGRQTW EFDPNAGTPQ ELAKVEEARR KFTEDRHTVK PASDLLWMMQ FMREKNFKQT IPPVRLGEE EQVTYEDLTT ALTRTTNFFT ALQASDGHWP AENGGVSFFL PPFIFSLYIT GHLNSIITPE YRKEILRFIY N HQNEDGGW ...String: MWKLKVAERG NAPYSEYLYT TNDFSGRQTW EFDPNAGTPQ ELAKVEEARR KFTEDRHTVK PASDLLWMMQ FMREKNFKQT IPPVRLGEE EQVTYEDLTT ALTRTTNFFT ALQASDGHWP AENGGVSFFL PPFIFSLYIT GHLNSIITPE YRKEILRFIY N HQNEDGGW GIHIEGHSTM FGTAFSYVCL RILGIEVDGG KDNACARARK WILDHGGITY MPSWGKTWLS ILGVYDWYGC NP MPPEFWL LPSYLPIHPA KIWCYCRMVY MPMSYLYGKR FVGPITPLIL QLREELHTQP FHEIQWRQTR HRCAKEDLYY PHS LIQDFI WDSLYVASEP LLTRWPLNKI REKALAKAME HIHYEDENSR YITIGCVEKA LCMLCCWVED PNSDYFKKHL ARIP DYLWV AEDGMKVQSF GSQLWDATFG FQALVASNLT EDEVGPALAK AYDFIKKSQV KDNPSGDFES MHRHISKGSW TFSDQ DHGW QLSDCTAEAL KCCLLAATMP QEVVGEKMKP EWVYEAINII LSLQSKSGGL AGWEPVRAGE WMEILNPMEF LENIVI EHT YVECTGSSII AFVSLKKLYP GHRTKDIDNF IRNAIRYLED VQYPDGSWYG NWGICFIYST MFALGGLAAT GRTYDNC QA VRRGVDFILK NQSDDGGWGE SYLSCPRKVY TPLDGRRSNV VQTAWAMLGL LYAGQAERDP TPLHRGAKVL INYQMEDG G YPQQEITGVF KMNCMLHYPI YRNAFPIWAL GEYRKRVPLP SKGY UniProtKB: Terpene cyclase/mutase family member |
-Macromolecule #2: octyl beta-D-glucopyranoside
| Macromolecule | Name: octyl beta-D-glucopyranoside / type: ligand / ID: 2 / Number of copies: 4 / Formula: BOG |
|---|---|
| Molecular weight | Theoretical: 292.369 Da |
| Chemical component information |  ChemComp-BOG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 2 mM dithiothreitol, and 0.2% OG |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.75 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 32698 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)