[English] 日本語
 Yorodumi
Yorodumi- EMDB-35950: The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constituti... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The truncated rice Na+/H+ antiporter SOS1 (1-976) in a constitutively active state | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Na+/H+ antiporter / cation:proton antiporter family 1 / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationpotassium:proton antiporter activity / sodium:proton antiporter activity / sodium ion import across plasma membrane / potassium ion transmembrane transport / regulation of intracellular pH / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
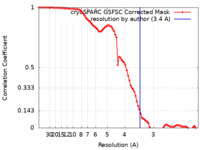
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhang XY / Tang LH / Zhang CR / Nie JW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na/H antiporter. Authors: Xiang-Yun Zhang / Ling-Hui Tang / Jia-Wei Nie / Chun-Rui Zhang / Xiaonan Han / Qi-Yu Li / Li Qin / Mei-Hua Wang / Xiahe Huang / Feifei Yu / Min Su / Yingchun Wang / Rui-Ming Xu / Yan Guo / ...Authors: Xiang-Yun Zhang / Ling-Hui Tang / Jia-Wei Nie / Chun-Rui Zhang / Xiaonan Han / Qi-Yu Li / Li Qin / Mei-Hua Wang / Xiahe Huang / Feifei Yu / Min Su / Yingchun Wang / Rui-Ming Xu / Yan Guo / Qi Xie / Yu-Hang Chen /  Abstract: Salinity is one of the most severe abiotic stresses that adversely affect plant growth and agricultural productivity. The plant Na/H antiporter Salt Overly Sensitive 1 (SOS1) located in the plasma ...Salinity is one of the most severe abiotic stresses that adversely affect plant growth and agricultural productivity. The plant Na/H antiporter Salt Overly Sensitive 1 (SOS1) located in the plasma membrane extrudes excess Na out of cells in response to salt stress and confers salt tolerance. However, the molecular mechanism underlying SOS1 activation remains largely elusive. Here we elucidate two cryo-electron microscopy structures of rice (Oryza sativa) SOS1, a full-length protein in an auto-inhibited state and a truncated version in an active state. The SOS1 forms a dimeric architecture, with an NhaA-folded transmembrane domain portion in the membrane and an elongated cytosolic portion of multiple regulatory domains in the cytoplasm. The structural comparison shows that SOS1 adopts an elevator transport mechanism accompanied by a conformational transition of the highly conserved Pro in the unwound transmembrane helix 5 (TM), switching from an occluded conformation in the auto-inhibited state to a conducting conformation in the active state. These findings allow us to propose an inhibition-release mechanism for SOS1 activation and elucidate how SOS1 controls Na homeostasis in response to salt stress. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35950.map.gz emd_35950.map.gz | 31.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35950-v30.xml emd-35950-v30.xml emd-35950.xml emd-35950.xml | 19.3 KB 19.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35950_fsc.xml emd_35950_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_35950.png emd_35950.png | 64.8 KB | ||
| Filedesc metadata |  emd-35950.cif.gz emd-35950.cif.gz | 6.5 KB | ||
| Others |  emd_35950_additional_1.map.gz emd_35950_additional_1.map.gz emd_35950_half_map_1.map.gz emd_35950_half_map_1.map.gz emd_35950_half_map_2.map.gz emd_35950_half_map_2.map.gz | 59.5 MB 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35950 http://ftp.pdbj.org/pub/emdb/structures/EMD-35950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35950 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35950 | HTTPS FTP |
-Validation report
| Summary document |  emd_35950_validation.pdf.gz emd_35950_validation.pdf.gz | 892.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35950_full_validation.pdf.gz emd_35950_full_validation.pdf.gz | 891.8 KB | Display | |
| Data in XML |  emd_35950_validation.xml.gz emd_35950_validation.xml.gz | 16.4 KB | Display | |
| Data in CIF |  emd_35950_validation.cif.gz emd_35950_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35950 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35950 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35950 | HTTPS FTP |
-Related structure data
| Related structure data |  8j2mMC  8iwoC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35950.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: sharpen map
| File | emd_35950_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpen map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35950_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35950_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : OsSOS1-1t
| Entire | Name: OsSOS1-1t |
|---|---|
| Components |
|
-Supramolecule #1: OsSOS1-1t
| Supramolecule | Name: OsSOS1-1t / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Na+/H+ antiporter
| Macromolecule | Name: Na+/H+ antiporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 116.560547 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDNPEAEPDD AVLFVGVSLV LGIASRHLLR GTRVPYTVAL LVLGVALGSL EFGTKHGMGK LGAGIRIWAN INPDLLLAVF LPALLFESS FSMEIHQIKK CMAQMVLLAG PGVLISTFFL GSALKLTFPY NWNWKTSLLL GGLLSATDPV AVVALLKELG A SKKLSTII ...String: MDNPEAEPDD AVLFVGVSLV LGIASRHLLR GTRVPYTVAL LVLGVALGSL EFGTKHGMGK LGAGIRIWAN INPDLLLAVF LPALLFESS FSMEIHQIKK CMAQMVLLAG PGVLISTFFL GSALKLTFPY NWNWKTSLLL GGLLSATDPV AVVALLKELG A SKKLSTII EGESLMNDGT AIVVYQLFYR MVLGRTFDAG SIIKFLSEVS LGAVALGLAF GIASVLWLGF IFNDTIIEIA LT LAVSYIA FFTAQDALEV SGVLTVMTLG MFYAAFAKTA FKGDSQQSLH HFWEMVAYIA NTLIFILSGV VIADGVLENN VHF ERHGAS WGFLLLLYVF VQISRILVVV ILYPLLRHFG YGLDLKEATI LVWAGLRGAV ALSLSLSVKR ASDAVQTHLK PVDG TMFVF FTGGIVFLTL IFNGSTTQFL LHLLGMDRLA ATKLRILNYT KYEMLNKALE AFGDLRDDEE LGPPADWVTV KKYIT CLND LDDEPVHPHA VSDRNDRMHT MNLRDIRVRL LNGVQAAYWG MLEEGRITQT TANILMRSVD EAMDLVPTQE LCDWKG LRS NVHFPNYYRF LQMSRLPRRL ITYFTVERLE SGCYICAAFL RAHRIARRQL HDFLGDSEVA RIVIDESNAE GEEARKF LE DVRVTFPQVL RVLKTRQVTY SVLTHLSEYI QNLQKTGLLE EKEMAHLDDA LQTDLKKFKR NPPLVKMPRV SDLLNTHP L VGALPAAMRD PLLSSTKETV KGHGTILYRE GSRPTGIWLV SIGVVKWTSQ RLSSRHSLDP ILSHGSTLGL YEVLIGKPY ICDMITDSVV HCFFIEAEKI EQLRQSDPSI EIFLWQESAL VVARLLLPMM FEKMATHELR VLITERSTMN IYIKGEEIEL EQNFIGILL EGFLKTKNQT LITPPGLLLP PNADLNLFGL ESSAINRIDY CYTAPSYQVE ARARILFVEI GRPEIEADLQ R SASLISQT LELPRTQSAA AENLYFQGLE WSHPQFEKGS GDYKDDDDKG SGWSHPQFEK LEDYKDDDDK HHHHHHHHHH UniProtKB: Na+/H+ antiporter |
-Macromolecule #2: (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradeca...
| Macromolecule | Name: (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl tetradecanoate type: ligand / ID: 2 / Number of copies: 2 / Formula: 46E |
|---|---|
| Molecular weight | Theoretical: 635.853 Da |
| Chemical component information |  ChemComp-46E: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.7 mg/mL |
|---|---|
| Buffer | pH: 6.5 |
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)