+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the AsCas12f-YHAM-sgRNAS3-5v7-target DNA | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / RNA BINDING PROTEIN-DNA COMPLEX / RNA BINDING PROTEIN / RNA BINDING PROTEIN-DNA-RNA complex | |||||||||
| Function / homology | : / Transposase IS605, OrfB, C-terminal / Putative transposase DNA-binding domain / endonuclease activity / Hydrolases; Acting on ester bonds / DNA binding / RNA binding / metal ion binding / CRISPR-associated endodeoxyribonuclease Cas12f1 Function and homology information Function and homology information | |||||||||
| Biological species |  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) | |||||||||
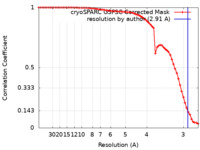
| Method | single particle reconstruction / cryo EM / Resolution: 2.91 Å | |||||||||
 Authors Authors | Hino T / Omura NS / Nakagawa R / Togashi T / Takeda NS / Hiramoto T / Tasaka S / Hirano H / Tokuyama T / Uosaki H ...Hino T / Omura NS / Nakagawa R / Togashi T / Takeda NS / Hiramoto T / Tasaka S / Hirano H / Tokuyama T / Uosaki H / Ishiguro H / Yamano H / Ozaki Y / Motooka D / Mori H / Kirita Y / Kise Y / Itoh Y / Matoba S / Aburatani H / Yachie N / Siksnys V / Ohmori T / Hoshino A / Nureki O | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: An AsCas12f-based compact genome-editing tool derived by deep mutational scanning and structural analysis. Authors: Tomohiro Hino / Satoshi N Omura / Ryoya Nakagawa / Tomoki Togashi / Satoru N Takeda / Takafumi Hiramoto / Satoshi Tasaka / Hisato Hirano / Takeshi Tokuyama / Hideki Uosaki / Soh Ishiguro / ...Authors: Tomohiro Hino / Satoshi N Omura / Ryoya Nakagawa / Tomoki Togashi / Satoru N Takeda / Takafumi Hiramoto / Satoshi Tasaka / Hisato Hirano / Takeshi Tokuyama / Hideki Uosaki / Soh Ishiguro / Madina Kagieva / Hiroyuki Yamano / Yuki Ozaki / Daisuke Motooka / Hideto Mori / Yuhei Kirita / Yoshiaki Kise / Yuzuru Itoh / Satoaki Matoba / Hiroyuki Aburatani / Nozomu Yachie / Tautvydas Karvelis / Virginijus Siksnys / Tsukasa Ohmori / Atsushi Hoshino / Osamu Nureki /   Abstract: SpCas9 and AsCas12a are widely utilized as genome-editing tools in human cells. However, their relatively large size poses a limitation for delivery by cargo-size-limited adeno-associated virus (AAV) ...SpCas9 and AsCas12a are widely utilized as genome-editing tools in human cells. However, their relatively large size poses a limitation for delivery by cargo-size-limited adeno-associated virus (AAV) vectors. The type V-F Cas12f from Acidibacillus sulfuroxidans is exceptionally compact (422 amino acids) and has been harnessed as a compact genome-editing tool. Here, we developed an approach, combining deep mutational scanning and structure-informed design, to successfully generate two AsCas12f activity-enhanced (enAsCas12f) variants. Remarkably, the enAsCas12f variants exhibited genome-editing activities in human cells comparable with those of SpCas9 and AsCas12a. The cryoelectron microscopy (cryo-EM) structures revealed that the mutations stabilize the dimer formation and reinforce interactions with nucleic acids to enhance their DNA cleavage activities. Moreover, enAsCas12f packaged with partner genes in an all-in-one AAV vector exhibited efficient knock-in/knock-out activities and transcriptional activation in mice. Taken together, enAsCas12f variants could offer a minimal genome-editing platform for in vivo gene therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35926.map.gz emd_35926.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35926-v30.xml emd-35926-v30.xml emd-35926.xml emd-35926.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35926_fsc.xml emd_35926_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35926.png emd_35926.png | 140.8 KB | ||
| Filedesc metadata |  emd-35926.cif.gz emd-35926.cif.gz | 6.4 KB | ||
| Others |  emd_35926_half_map_1.map.gz emd_35926_half_map_1.map.gz emd_35926_half_map_2.map.gz emd_35926_half_map_2.map.gz | 4.5 MB 4.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35926 http://ftp.pdbj.org/pub/emdb/structures/EMD-35926 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35926 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35926 | HTTPS FTP |
-Validation report
| Summary document |  emd_35926_validation.pdf.gz emd_35926_validation.pdf.gz | 711.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35926_full_validation.pdf.gz emd_35926_full_validation.pdf.gz | 711.1 KB | Display | |
| Data in XML |  emd_35926_validation.xml.gz emd_35926_validation.xml.gz | 11.2 KB | Display | |
| Data in CIF |  emd_35926_validation.cif.gz emd_35926_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35926 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35926 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35926 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35926 | HTTPS FTP |
-Related structure data
| Related structure data |  8j1jMC  8j12C  8j3rC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35926.map.gz / Format: CCP4 / Size: 4.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35926.map.gz / Format: CCP4 / Size: 4.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.328 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35926_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35926_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : AsCas12f-YHAM-sgRNAS3-5v7-target DNA
| Entire | Name: AsCas12f-YHAM-sgRNAS3-5v7-target DNA |
|---|---|
| Components |
|
-Supramolecule #1: AsCas12f-YHAM-sgRNAS3-5v7-target DNA
| Supramolecule | Name: AsCas12f-YHAM-sgRNAS3-5v7-target DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) |
-Macromolecule #1: Transposase IS605 OrfB C-terminal domain-containing protein
| Macromolecule | Name: Transposase IS605 OrfB C-terminal domain-containing protein type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) |
| Molecular weight | Theoretical: 49.966105 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHGS MIKVYRYEIV KPLDLDWKEF GTILRQLQQE TRFALNKATQ LAWEWMGYSS DYKDNHGEYP KSKDILGYTN VHGYAYHTI KTKAYRLNSG NLSQTIKRAT DRFKAYQKEI LRGDMSIPSY KRDIPLDLIK ENISVNRMNH GDYIASLSLL S NPAKQEMN ...String: MGHHHHHHGS MIKVYRYEIV KPLDLDWKEF GTILRQLQQE TRFALNKATQ LAWEWMGYSS DYKDNHGEYP KSKDILGYTN VHGYAYHTI KTKAYRLNSG NLSQTIKRAT DRFKAYQKEI LRGDMSIPSY KRDIPLDLIK ENISVNRMNH GDYIASLSLL S NPAKQEMN VKRKISVIII VRGAGKTIMD RILSGEYQVH ASQIIHDDRK NKWYLNISYD FEPQTRVLDL NKIMGIDLGV AV AAYMAFQ HTPARYKLEG GEIENFRRQV ESRRISMLRQ GKYAGGARGG HGRDKRIKPI EQLRDKIANF RDTTNHRYSR YIV DMAIKM GCGTIQMEDL TNIRDIGSRF LQNWTYYDLQ QKIIYKAEEA GIKVIKIDPQ YTSQRCSECG NIDSGNRIGQ AIFK CRACG YEANADYNAA RNIAIPNIDK IIAESIK UniProtKB: CRISPR-associated endodeoxyribonuclease Cas12f1 |
-Macromolecule #2: DNA (38-MER)
| Macromolecule | Name: DNA (38-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) |
| Molecular weight | Theoretical: 11.694576 KDa |
| Sequence | String: (DG)(DA)(DA)(DT)(DG)(DG)(DT)(DT)(DC)(DA) (DA)(DG)(DC)(DG)(DC)(DA)(DC)(DC)(DT)(DA) (DA)(DT)(DT)(DT)(DC)(DC)(DT)(DA)(DA) (DA)(DT)(DT)(DA)(DG)(DA)(DA)(DA)(DA) |
-Macromolecule #3: DNA (38-MER)
| Macromolecule | Name: DNA (38-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) |
| Molecular weight | Theoretical: 11.689535 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DC)(DT)(DA)(DA)(DT)(DT) (DT)(DA)(DG)(DG)(DA)(DA)(DA)(DT)(DT)(DA) (DG)(DG)(DT)(DG)(DC)(DG)(DC)(DT)(DT) (DG)(DA)(DA)(DC)(DC)(DA)(DT)(DT)(DC) |
-Macromolecule #4: RNA (118-MER)
| Macromolecule | Name: RNA (118-MER) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Sulfoacidibacillus thermotolerans (bacteria) Sulfoacidibacillus thermotolerans (bacteria) |
| Molecular weight | Theoretical: 38.288805 KDa |
| Sequence | String: GGAUUCGUCG GUUCAGCGAC GAUAAGCCGA GAAGUGCCAA UAAAACUGUU AAGUGGUUUG GUAACGCUCG GUAAGGUCCG AAAGGAGAA CCACUGAACG GAAAUUAGGU GCGCUUGGC |
-Macromolecule #5: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 5 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #6: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 6 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)