| 登録情報 | データベース: EMDB / ID: EMD-35866
|
|---|
| タイトル | PKR and NS1 complex |
|---|
 マップデータ マップデータ | |
|---|
 試料 試料 | - 複合体: PKR and NS1 complex
- タンパク質・ペプチド: Interferon-induced, double-stranded RNA-activated protein kinase
- タンパク質・ペプチド: Non-structural protein 1
|
|---|
 キーワード キーワード | PKR / NS1 / PKR and NS1 complex / VIRUS / TRANSFERASE-VIRAL PROTEIN complex |
|---|
| 機能・相同性 |  機能・相同性情報 機能・相同性情報
Inhibition of PKR / regulation of NLRP3 inflammasome complex assembly / symbiont-mediated suppression of host mRNA processing / response to interferon-alpha / negative regulation of osteoblast proliferation / symbiont-mediated suppression of host PKR/eIFalpha signaling / regulation of hematopoietic progenitor cell differentiation / positive regulation of stress-activated MAPK cascade / protein phosphatase regulator activity / SUMOylation of immune response proteins ...Inhibition of PKR / regulation of NLRP3 inflammasome complex assembly / symbiont-mediated suppression of host mRNA processing / response to interferon-alpha / negative regulation of osteoblast proliferation / symbiont-mediated suppression of host PKR/eIFalpha signaling / regulation of hematopoietic progenitor cell differentiation / positive regulation of stress-activated MAPK cascade / protein phosphatase regulator activity / SUMOylation of immune response proteins / regulation of hematopoietic stem cell proliferation / protein serine/threonine kinase inhibitor activity / regulation of translational initiation / regulation of hematopoietic stem cell differentiation / negative regulation of viral genome replication / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / endoplasmic reticulum unfolded protein response / positive regulation of chemokine production / antiviral innate immune response / cellular response to amino acid starvation / positive regulation of cytokine production / non-membrane spanning protein tyrosine kinase activity / non-specific protein-tyrosine kinase / positive regulation of non-canonical NF-kappaB signal transduction / PKR-mediated signaling / response to virus / Evasion by RSV of host interferon responses / ISG15 antiviral mechanism / Interferon alpha/beta signaling / kinase activity / positive regulation of NF-kappaB transcription factor activity / double-stranded RNA binding / protein autophosphorylation / defense response to virus / eukaryotic translation initiation factor 2alpha kinase activity / host cell cytoplasm / positive regulation of MAPK cascade / 3-phosphoinositide-dependent protein kinase activity / DNA-dependent protein kinase activity / ribosomal protein S6 kinase activity / histone H3S10 kinase activity / histone H2AXS139 kinase activity / histone H3S28 kinase activity / histone H4S1 kinase activity / histone H2BS14 kinase activity / histone H3T3 kinase activity / histone H2AS121 kinase activity / Rho-dependent protein serine/threonine kinase activity / histone H2BS36 kinase activity / histone H3S57 kinase activity / histone H2AT120 kinase activity / AMP-activated protein kinase activity / histone H2AS1 kinase activity / histone H3T6 kinase activity / histone H3T11 kinase activity / histone H3T45 kinase activity / non-specific serine/threonine protein kinase / protein kinase activity / negative regulation of translation / : / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / ribosome / protein phosphorylation / translation / symbiont-mediated suppression of host gene expression / negative regulation of cell population proliferation / protein serine kinase activity / protein serine/threonine kinase activity / negative regulation of apoptotic process / host cell nucleus / perinuclear region of cytoplasm / RNA binding / nucleoplasm / ATP binding / identical protein binding / nucleus / membrane / cytosol / cytoplasm類似検索 - 分子機能 EIF2AK2, first double-stranded RNA binding domain / EIF2AK2, second double-stranded RNA binding domain / Influenza A virus NS1 protein / Influenza A virus NS1, effector domain-like superfamily / Influenza non-structural protein (NS1) / Influenza non-structural protein (NS1) / : / Double-stranded RNA binding motif / Double-stranded RNA binding motif / Double stranded RNA-binding domain (dsRBD) profile. ...EIF2AK2, first double-stranded RNA binding domain / EIF2AK2, second double-stranded RNA binding domain / Influenza A virus NS1 protein / Influenza A virus NS1, effector domain-like superfamily / Influenza non-structural protein (NS1) / Influenza non-structural protein (NS1) / : / Double-stranded RNA binding motif / Double-stranded RNA binding motif / Double stranded RNA-binding domain (dsRBD) profile. / Double-stranded RNA-binding domain / S15/NS1, RNA-binding / Serine/threonine-protein kinase, active site / Serine/Threonine protein kinases active-site signature. / Protein kinase domain / Serine/Threonine protein kinases, catalytic domain / Protein kinase, ATP binding site / Protein kinases ATP-binding region signature. / Protein kinase domain profile. / Protein kinase domain / Protein kinase-like domain superfamily類似検索 - ドメイン・相同性 Interferon-induced, double-stranded RNA-activated protein kinase / Non-structural protein 1類似検索 - 構成要素 |
|---|
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス) Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス) |
|---|
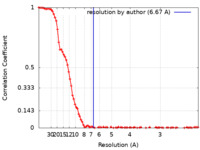
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 6.67 Å |
|---|
 データ登録者 データ登録者 | Han CW / Kim HJ |
|---|
| 資金援助 | 1件 | Organization | Grant number | 国 |
|---|
| Not funded | | |
|
|---|
 引用 引用 |  ジャーナル: To Be Published ジャーナル: To Be Published
タイトル: Structure of PKR and NS1 complex
著者: Han CW |
|---|
| 履歴 | | 登録 | 2023年4月7日 | - |
|---|
| ヘッダ(付随情報) 公開 | 2024年4月10日 | - |
|---|
| マップ公開 | 2024年4月10日 | - |
|---|
| 更新 | 2024年4月10日 | - |
|---|
| 現状 | 2024年4月10日 | 処理サイト: PDBj / 状態: 公開 |
|---|
|
|---|
 データを開く
データを開く 基本情報
基本情報
 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト) /
Homo sapiens (ヒト) /  Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス)
Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス) データ登録者
データ登録者 引用
引用 ジャーナル: To Be Published
ジャーナル: To Be Published 構造の表示
構造の表示 ダウンロードとリンク
ダウンロードとリンク emd_35866.map.gz
emd_35866.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-35866-v30.xml
emd-35866-v30.xml emd-35866.xml
emd-35866.xml EMDBヘッダ
EMDBヘッダ emd_35866_fsc.xml
emd_35866_fsc.xml FSCデータファイル
FSCデータファイル emd_35866.png
emd_35866.png emd-35866.cif.gz
emd-35866.cif.gz emd_35866_additional_1.map.gz
emd_35866_additional_1.map.gz emd_35866_half_map_1.map.gz
emd_35866_half_map_1.map.gz emd_35866_half_map_2.map.gz
emd_35866_half_map_2.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-35866
http://ftp.pdbj.org/pub/emdb/structures/EMD-35866 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35866
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35866 emd_35866_validation.pdf.gz
emd_35866_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_35866_full_validation.pdf.gz
emd_35866_full_validation.pdf.gz emd_35866_validation.xml.gz
emd_35866_validation.xml.gz emd_35866_validation.cif.gz
emd_35866_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35866
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35866 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35866
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35866
 F&H 検索
F&H 検索 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_35866.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_35866.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素 Homo sapiens (ヒト)
Homo sapiens (ヒト) Homo sapiens (ヒト)
Homo sapiens (ヒト)
 Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス)
Influenza A virus (strain A/Memphis/102/1972 H3N2) (A型インフルエンザウイルス)
 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN ムービー
ムービー コントローラー
コントローラー






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)