[English] 日本語
 Yorodumi
Yorodumi- EMDB-35832: Cryo-EM structure of the GPR34 receptor in complex with the antag... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Inhibitor / GPR34 receptor in complex with the antagonist YL-365 / MEMBRANE PROTEIN / MEMBRANE PROTEIN-INHIBITOR complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled purinergic nucleotide receptor activity / G protein-coupled receptor activity / G protein-coupled receptor signaling pathway / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
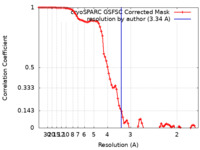
| Method | single particle reconstruction / cryo EM / Resolution: 3.34 Å | ||||||||||||
 Authors Authors | Jia GW / Wang X / Zhang CB / Dong HH / Su ZM | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Cryo-EM structures of human GPR34 enable the identification of selective antagonists. Authors: Anjie Xia / Xihao Yong / Changbin Zhang / Guifeng Lin / Guowen Jia / Chang Zhao / Xin Wang / Yize Hao / Yifei Wang / Pei Zhou / Xin Yang / Yue Deng / Chao Wu / Yujiao Chen / Jiawei Zhu / ...Authors: Anjie Xia / Xihao Yong / Changbin Zhang / Guifeng Lin / Guowen Jia / Chang Zhao / Xin Wang / Yize Hao / Yifei Wang / Pei Zhou / Xin Yang / Yue Deng / Chao Wu / Yujiao Chen / Jiawei Zhu / Xiaodi Tang / Jingming Liu / Shiyu Zhang / Jiahao Zhang / Zheng Xu / Qian Hu / Jinlong Zhao / Yuting Yue / Wei Yan / Zhaoming Su / Yuquan Wei / Rongbin Zhou / Haohao Dong / Zhenhua Shao / Shengyong Yang /  Abstract: GPR34 is a functional G-protein-coupled receptor of Lysophosphatidylserine (LysoPS), and has pathogenic roles in numerous diseases, yet remains poorly targeted. We herein report a cryo-electron ...GPR34 is a functional G-protein-coupled receptor of Lysophosphatidylserine (LysoPS), and has pathogenic roles in numerous diseases, yet remains poorly targeted. We herein report a cryo-electron microscopy (cryo-EM) structure of GPR34 bound with LysoPS (18:1) and G protein, revealing a unique ligand recognition mode with the negatively charged head group of LysoPS occupying a polar cavity formed by TM3, 6 and 7, and the hydrophobic tail of LysoPS residing in a lateral open hydrophobic groove formed by TM3-5. Virtual screening and subsequent structural optimization led to the identification of a highly potent and selective antagonist (YL-365). Design of fusion proteins allowed successful determination of the challenging cryo-EM structure of the inactive GPR34 complexed with YL-365, which revealed the competitive binding of YL-365 in a portion of the orthosteric binding pocket of GPR34 and the antagonist-binding-induced allostery in the receptor, implicating the inhibition mechanism of YL-365. Moreover, YL-365 displayed excellent activity in a neuropathic pain model without obvious toxicity. Collectively, this study offers mechanistic insights into the endogenous agonist recognition and antagonist inhibition of GPR34, and provides proof of concept that targeting GPR34 represents a promising strategy for disease treatment. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35832.map.gz emd_35832.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35832-v30.xml emd-35832-v30.xml emd-35832.xml emd-35832.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35832_fsc.xml emd_35832_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35832.png emd_35832.png | 84.9 KB | ||
| Filedesc metadata |  emd-35832.cif.gz emd-35832.cif.gz | 5.9 KB | ||
| Others |  emd_35832_half_map_1.map.gz emd_35832_half_map_1.map.gz emd_35832_half_map_2.map.gz emd_35832_half_map_2.map.gz | 59.1 MB 59.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35832 http://ftp.pdbj.org/pub/emdb/structures/EMD-35832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35832 | HTTPS FTP |
-Validation report
| Summary document |  emd_35832_validation.pdf.gz emd_35832_validation.pdf.gz | 808.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35832_full_validation.pdf.gz emd_35832_full_validation.pdf.gz | 808.3 KB | Display | |
| Data in XML |  emd_35832_validation.xml.gz emd_35832_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_35832_validation.cif.gz emd_35832_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35832 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35832 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35832 | HTTPS FTP |
-Related structure data
| Related structure data |  8iyxMC  8saiC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35832.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35832.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35832_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35832_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the GPR34 receptor in complex with the antag...
| Entire | Name: Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the GPR34 receptor in complex with the antag...
| Supramolecule | Name: Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Probable G-protein coupled receptor 34,Probable G-protein coupled...
| Macromolecule | Name: Probable G-protein coupled receptor 34,Probable G-protein coupled receptor 34,YL-365 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.231812 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRSHTITMTT TSVSSWPYSS HRMRFITNHS DQPPQNFSAT PNVTTCPMDE KLLSTVLTTS YSVIFIVGLV GNIIALYVFL GIHRKRNSI QIYLLNVAIA DLLLIFCLPF RIMYHINQNK WTLGVILCKV VGTLFYMNMY ISIILLGFIS LDRYIKINRS I QQRKAITT ...String: MRSHTITMTT TSVSSWPYSS HRMRFITNHS DQPPQNFSAT PNVTTCPMDE KLLSTVLTTS YSVIFIVGLV GNIIALYVFL GIHRKRNSI QIYLLNVAIA DLLLIFCLPF RIMYHINQNK WTLGVILCKV VGTLFYMNMY ISIILLGFIS LDRYIKINRS I QQRKAITT KQSIYVCCIV WMLALGGFLT MIILTLKKGG HNSTMCFHYR DKHNAKGEAI FNFILVVMFW LIFLLIILSY IK IGKNLLR ISKRGIDCSF WNESYLTGSR DERKKSLLSK FGMDEGVTFM FIGRFDRGQK GVDVLLKAIE ILSSKKEFQE MRF IIIGKG DPELEGWARS LEEKHGNVKV ITEMLSREFV RELYGSVDFV IIPSYFEPFG LVALEAMCLG AIPIASAVGG LRDI ITNET GILVKAGDPG ELANAILKAL ELSRSDLSKF RENCKKRAMS FSDQARMDIR LAKGKYATTA RNSFIVLIIF TICFV PYHA FRFIYISSQL NVSSCYWKEI VHKTNEIMLV LSSFNSCLDP VMYFLMSSNI RKIMCQLLFR RFQGEPSRSE STSEFK PGY SLHDTSVAVK IQSSSKST UniProtKB: Probable G-protein coupled receptor 34, Probable G-protein coupled receptor 34 |
-Macromolecule #2: 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)...
| Macromolecule | Name: 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid type: ligand / ID: 2 / Number of copies: 1 / Formula: S6R |
|---|---|
| Molecular weight | Theoretical: 583.073 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 56.7 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)