[English] 日本語
 Yorodumi
Yorodumi- EMDB-35784: Cryo-EM structure of unprotonated LHCII in detergent solution at ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of unprotonated LHCII in detergent solution at low pH value | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | LHCII / Light-harvesting complex / Photosystem II / PLANT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationphotosynthesis, light harvesting / photosystem I / photosystem II / chloroplast thylakoid membrane / chlorophyll binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Spinacia oleracea (spinach) Spinacia oleracea (spinach) | |||||||||
| Method | single particle reconstruction / Resolution: 2.52 Å | |||||||||
 Authors Authors | Ruan MX / Ding W | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Plants / Year: 2023 Journal: Nat Plants / Year: 2023Title: Cryo-EM structures of LHCII in photo-active and photo-protecting states reveal allosteric regulation of light harvesting and excess energy dissipation. Authors: Meixia Ruan / Hao Li / Ying Zhang / Ruoqi Zhao / Jun Zhang / Yingjie Wang / Jiali Gao / Zhuan Wang / Yumei Wang / Dapeng Sun / Wei Ding / Yuxiang Weng /   Abstract: The major light-harvesting complex of photosystem II (LHCII) has a dual regulatory function in a process called non-photochemical quenching to avoid the formation of reactive oxygen. LHCII undergoes ...The major light-harvesting complex of photosystem II (LHCII) has a dual regulatory function in a process called non-photochemical quenching to avoid the formation of reactive oxygen. LHCII undergoes reversible conformation transitions to switch between a light-harvesting state for excited-state energy transfer and an energy-quenching state for dissipating excess energy under full sunshine. Here we report cryo-electron microscopy structures of LHCII in membrane nanodiscs, which mimic in vivo LHCII, and in detergent solution at pH 7.8 and 5.4, respectively. We found that, under low pH conditions, the salt bridges at the lumenal side of LHCII are broken, accompanied by the formation of two local α-helices on the lumen side. The formation of α-helices in turn triggers allosterically global protein conformational change, resulting in a smaller crossing angle between transmembrane helices. The fluorescence decay rates corresponding to different conformational states follow the Dexter energy transfer mechanism with a characteristic transition distance of 5.6 Å between Lut1 and Chl612. The experimental observations are consistent with the computed electronic coupling strengths using multistate density function theory. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35784.map.gz emd_35784.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35784-v30.xml emd-35784-v30.xml emd-35784.xml emd-35784.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35784.png emd_35784.png | 145.8 KB | ||
| Filedesc metadata |  emd-35784.cif.gz emd-35784.cif.gz | 6.1 KB | ||
| Others |  emd_35784_half_map_1.map.gz emd_35784_half_map_1.map.gz emd_35784_half_map_2.map.gz emd_35784_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35784 http://ftp.pdbj.org/pub/emdb/structures/EMD-35784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35784 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35784 | HTTPS FTP |
-Validation report
| Summary document |  emd_35784_validation.pdf.gz emd_35784_validation.pdf.gz | 819.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35784_full_validation.pdf.gz emd_35784_full_validation.pdf.gz | 819.2 KB | Display | |
| Data in XML |  emd_35784_validation.xml.gz emd_35784_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_35784_validation.cif.gz emd_35784_validation.cif.gz | 14.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35784 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35784 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35784 | HTTPS FTP |
-Related structure data
| Related structure data |  8iwzMC  8iwxC  8iwyC  8ix0C  8ix1C  8ix2C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35784.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35784.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
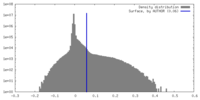
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35784_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35784_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : LHCII in detergent solution at low pH value
| Entire | Name: LHCII in detergent solution at low pH value |
|---|---|
| Components |
|
-Supramolecule #1: LHCII in detergent solution at low pH value
| Supramolecule | Name: LHCII in detergent solution at low pH value / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Molecular weight | Theoretical: 120 KDa |
-Macromolecule #1: Chlorophyll a-b binding protein, chloroplastic
| Macromolecule | Name: Chlorophyll a-b binding protein, chloroplastic / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Molecular weight | Theoretical: 23.497549 KDa |
| Recombinant expression | Organism:  Spinacia oleracea (spinach) Spinacia oleracea (spinach) |
| Sequence | String: SPWYGPDRVK YLGPFSGESP SYLTGEFPGD YGWDTAGLSA DPETFAKNRE LEVIHCRWAM LGALGCVFPE LLARNGVKFG EAVWFKAGS QIFSEGGLDY LGNPSLVHAQ SILAIWACQV ILMGAVEGYR IAGGPLGEVV DPLYPGGSFD PLGLADDPEA F AELKVKEI ...String: SPWYGPDRVK YLGPFSGESP SYLTGEFPGD YGWDTAGLSA DPETFAKNRE LEVIHCRWAM LGALGCVFPE LLARNGVKFG EAVWFKAGS QIFSEGGLDY LGNPSLVHAQ SILAIWACQV ILMGAVEGYR IAGGPLGEVV DPLYPGGSFD PLGLADDPEA F AELKVKEI KNGRLAMFSM FGFFVQAIVT GKGPLENLAD HLADPVNNNA WNFATNFVPG UniProtKB: Chlorophyll a-b binding protein, chloroplastic |
-Macromolecule #2: CHLOROPHYLL B
| Macromolecule | Name: CHLOROPHYLL B / type: ligand / ID: 2 / Number of copies: 18 / Formula: CHL |
|---|---|
| Molecular weight | Theoretical: 907.472 Da |
| Chemical component information |  ChemComp-CHL: |
-Macromolecule #3: CHLOROPHYLL A
| Macromolecule | Name: CHLOROPHYLL A / type: ligand / ID: 3 / Number of copies: 24 / Formula: CLA |
|---|---|
| Molecular weight | Theoretical: 893.489 Da |
| Chemical component information |  ChemComp-CLA: |
-Macromolecule #4: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL
| Macromolecule | Name: (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL type: ligand / ID: 4 / Number of copies: 6 / Formula: LUT |
|---|---|
| Molecular weight | Theoretical: 568.871 Da |
| Chemical component information |  ChemComp-LUT: |
-Macromolecule #5: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY...
| Macromolecule | Name: (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL type: ligand / ID: 5 / Number of copies: 3 / Formula: NEX |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-NEX: |
-Macromolecule #6: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE
| Macromolecule | Name: 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE / type: ligand / ID: 6 / Number of copies: 3 / Formula: LHG |
|---|---|
| Molecular weight | Theoretical: 722.97 Da |
| Chemical component information |  ChemComp-LHG: |
-Macromolecule #7: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BE...
| Macromolecule | Name: (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL type: ligand / ID: 7 / Number of copies: 3 / Formula: XAT |
|---|---|
| Molecular weight | Theoretical: 600.87 Da |
| Chemical component information |  ChemComp-XAT: |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 5.4 / Component - Concentration: 10.0 mM / Component - Formula: Tris-HCL / Component - Name: Tris-HCL / Details: 10mM Tris-HCL, 0.03%beta-DDM |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 25 sec. / Pretreatment - Atmosphere: OTHER / Pretreatment - Pressure: 7.000000000000001e-05 kPa |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 80.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 7282 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.8 µm / Calibrated magnification: 22500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.8 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 2.52 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 879190 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)