+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
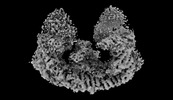
| Title | Cryo-EM structure of mouse BIRC6, Global map | |||||||||
 Map data Map data | Author stated: The map in this deposition is a global map in which the N-terminal section is relative floppy so that the atom inclusion of that section is very low. To address the bad atom inclusion, we have deposited the optimized local density map of the N-terminal section separately as EMD-35758. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords |  Inhibitor of apoptosis protein / BIRC6 / Smac / Inhibitor of apoptosis protein / BIRC6 / Smac /  APOPTOSIS APOPTOSIS | |||||||||
| Function / homology |  Function and homology information Function and homology informationspongiotrophoblast layer development / labyrinthine layer development / Flemming body /  microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / placenta development / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / placenta development /  regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase ...spongiotrophoblast layer development / labyrinthine layer development / Flemming body / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase ...spongiotrophoblast layer development / labyrinthine layer development / Flemming body /  microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / placenta development / microtubule organizing center / cysteine-type endopeptidase inhibitor activity / ubiquitin conjugating enzyme activity / placenta development /  regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase / regulation of cytokinesis / negative regulation of extrinsic apoptotic signaling pathway / RING-type E3 ubiquitin transferase /  trans-Golgi network / trans-Golgi network /  spindle pole / ubiquitin-protein transferase activity / regulation of cell population proliferation / midbody / cell population proliferation / protein ubiquitination / spindle pole / ubiquitin-protein transferase activity / regulation of cell population proliferation / midbody / cell population proliferation / protein ubiquitination /  endosome / endosome /  cell cycle / cell cycle /  cell division / apoptotic process / positive regulation of cell population proliferation / negative regulation of apoptotic process / cell division / apoptotic process / positive regulation of cell population proliferation / negative regulation of apoptotic process /  membrane / membrane /  nucleus nucleusSimilarity search - Function | |||||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.6 Å cryo EM / Resolution: 3.6 Å | |||||||||
 Authors Authors | Liu S / Jiang T / Bu F / Zhao J / Wang G / Li N / Gao N / Qiu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy. Authors: Shuo-Shuo Liu / Tian-Xia Jiang / Fan Bu / Ji-Lan Zhao / Guang-Fei Wang / Guo-Heng Yang / Jie-Yan Kong / Yun-Fan Qie / Pei Wen / Li-Bin Fan / Ning-Ning Li / Ning Gao / Xiao-Bo Qiu /  Abstract: Procaspase 9 is the initiator caspase for apoptosis, but how its levels and activities are maintained remains unclear. The gigantic Inhibitor-of-Apoptosis Protein BIRC6/BRUCE/Apollon inhibits both ...Procaspase 9 is the initiator caspase for apoptosis, but how its levels and activities are maintained remains unclear. The gigantic Inhibitor-of-Apoptosis Protein BIRC6/BRUCE/Apollon inhibits both apoptosis and autophagy by promoting ubiquitylation of proapoptotic factors and the key autophagic protein LC3, respectively. Here we show that BIRC6 forms an anti-parallel U-shaped dimer with multiple previously unannotated domains, including a ubiquitin-like domain, and the proapoptotic factor Smac/DIABLO binds BIRC6 in the central cavity. Notably, Smac outcompetes the effector caspase 3 and the pro-apoptotic protease HtrA2, but not procaspase 9, for binding BIRC6 in cells. BIRC6 also binds LC3 through its LC3-interacting region, probably following dimer disruption of this BIRC6 region. Mutation at LC3 ubiquitylation site promotes autophagy and autophagic degradation of BIRC6. Moreover, induction of autophagy promotes autophagic degradation of BIRC6 and caspase 9, but not of other effector caspases. These results are important to understand how the balance between apoptosis and autophagy is regulated under pathophysiological conditions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35759.map.gz emd_35759.map.gz | 11.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35759-v30.xml emd-35759-v30.xml emd-35759.xml emd-35759.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35759.png emd_35759.png | 43.3 KB | ||
| Masks |  emd_35759_msk_1.map emd_35759_msk_1.map | 125 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-35759.cif.gz emd-35759.cif.gz | 9 KB | ||
| Others |  emd_35759_half_map_1.map.gz emd_35759_half_map_1.map.gz emd_35759_half_map_2.map.gz emd_35759_half_map_2.map.gz | 97.2 MB 97.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35759 http://ftp.pdbj.org/pub/emdb/structures/EMD-35759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35759 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35759 | HTTPS FTP |
-Related structure data
| Related structure data |  8ivqMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35759.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35759.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Author stated: The map in this deposition is a global map in which the N-terminal section is relative floppy so that the atom inclusion of that section is very low. To address the bad atom inclusion, we have deposited the optimized local density map of the N-terminal section separately as EMD-35758. | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.37 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_35759_msk_1.map emd_35759_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_35759_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35759_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Baculoviral IAP repeat-containing protein 6, Dimer
| Entire | Name: Baculoviral IAP repeat-containing protein 6, Dimer |
|---|---|
| Components |
|
-Supramolecule #1: Baculoviral IAP repeat-containing protein 6, Dimer
| Supramolecule | Name: Baculoviral IAP repeat-containing protein 6, Dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
-Macromolecule #1: Isoform 2 of Baculoviral IAP repeat-containing protein 6
| Macromolecule | Name: Isoform 2 of Baculoviral IAP repeat-containing protein 6 type: protein_or_peptide / ID: 1 Details: The plasmid used for BIRC6 protein expression in this deposition is originated from Dr. Stefan Jentsch lab, and the Birc6 sequence in that plasmid is available from GenBank under accession ...Details: The plasmid used for BIRC6 protein expression in this deposition is originated from Dr. Stefan Jentsch lab, and the Birc6 sequence in that plasmid is available from GenBank under accession number GenBank: Y17267.1. No identical sequence could be identified in Uniprot. Here is the citation of the used Birc6 sequence: Hauser, H.P., et al., A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. Journal of Cell Biology, 1998. 141(6): p. 1415-1422. Number of copies: 2 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:   Mus musculus (house mouse) Mus musculus (house mouse) |
| Molecular weight | Theoretical: 535.020812 KDa |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKRVVPLES TGLQELATME QKLISEEDLE FMAAAAAEAS GPSCSSAAAA AGAGAAGVSE WLVLRDGCM RCDADGLHSL SYHPALNAIL AVTSRGTIKV IDGTSGATLQ ASALSAKPGG QVKCQYISAV DKVIFVDDYA V GCRKDLNG ...String: MDYKDHDGDY KDHDIDYKDD DDKRVVPLES TGLQELATME QKLISEEDLE FMAAAAAEAS GPSCSSAAAA AGAGAAGVSE WLVLRDGCM RCDADGLHSL SYHPALNAIL AVTSRGTIKV IDGTSGATLQ ASALSAKPGG QVKCQYISAV DKVIFVDDYA V GCRKDLNG ILLLDTALQT PVSKQDDVVQ LELPVTEAQQ LLSACIEKID VSSTEGYDLF ITQLKDGLKN ISHETAANHK VA KWATVTF HLPHHVLKSI ASAIVNELKK INQNVAALPV ASSVMDRLSY LLPSARPELG VGPGRSVDRA LMYSEANRRE TFT SWPHVG YRWAQPDPMA QAGFYHQPAS SGDDRAMCFT CSVCLVCWEP TDEPWSEHER HSPNCPFVKG EHTQNVPLSV TLAT SPAQL PSADGADRIA CFGSGSCPQF LAAATKRGKI CIWDVSKLMK VHLKFEINAY DPAIVQQLIL SGDPSSGVDS RRPTL AWLE DSSSCSDIPK LEGDSDDLLE DSDSEEHSRS DSVTGHTSQK EAMEVSLDIT ALSILQQPEK LQWEIVANVL EDTVKD LEE LGANPSLTNS KSEKTKEKHQ EQHNIPFPCL LAGGLLTYKS PATSPISSNS HRSLDGLSRT QGESISEQGS TDNESCT NS ELNSPLVRRT LPVLLLYSIK ESDEKAGKIF SQMNNIMSKS LHDDGFTVPQ IIEMELDNQE QLLLQDPPVT YIQQFADA A ASLTSPDSEK WNSVFPKPGT LVQCLRLPKF AEEETLCIDS ITPCADGIHL LVGLRTCSVE SLSAINQVEA LNNLNKLNS ALCNRRKGDL ESNLAVVNGA NISVIQHESP ADVPEHLLIR PEQRNVVSGG YLVLYKMNYT TRIVTLEEEP VKIQHIKDPQ DTITSLILL PPDILDNRED DCEEPAEEMQ LASKNGIERE KKSDISTLGH LVVTTQGGYV KVLDLSNFEI LAKVEPPKKE G TEEQDTFV SVIYCSGTDR LCACTKGGEL HFLQIGGTCD DIDEADILVD GSLSKGIEPA LEGSRPLSNP SSPGISGVEL LV DQPFTLE ILTSLVELTR FETLTPRFSA TVPPCWVEVQ QEQQQRRHPQ HLHQQHHGDA AQHTRTWKLQ TDSNSWDEHV FEL VLPKAC MVGHVDFKFV LNSNITSVPQ IQVTLLKNKA PGLGKANALN IEVEHNGNPS LVDLNEEMHH MDVEESQCLR LCPF LEDHK EDILCGPVWL ASGLDLSGHA GMLTLTSPKL VKGMAGGKYR SFLIHVKAVS DRGAADEMCS SGLRPVVRLP SLKQQ GHKG YSLASLLAKV AAGKEKSSNV KNENAGGTRK SENLRGCDLL QEVSVTIRRF KKTSICKERV QRCAMLQFSE FHEKLL NTL CRRSDDGQVT EHAQSLVLDA LCWLAGVHSN GSGSSKEGNE CLLSKTRKCL SDIVRVCFFE AGRSIAHKCA RFLALCI SN GKCEPCQPGF GSVLLKALLD NMCFLPAAAT GGSVYWYFVL LNYVKDEDLA GCSTACAALL TAVSRQLQDR LTPLEALL Q TRYGLYSSPF DPVLFDLEMS GSSWKTVYSS STAVQSDEID LSDVLSGNGR VSSCTAAEGS FTSLTGLLEV EPLHFTCVS TSDGTRIERD DASTFTVSSF GVPPAVGGLS SGTVGEASTA LSSAAQVALQ SLSHAMASAE QQLQVLQEKQ QQLLKLQQQK AKLEAKLHQ TTAAAAAAAS AAAAAAAGPV HNAVPSNPVA APGFFIHPSD VIPPTPKTTP LFMTPPLTPP NEAVSVVINA E LAQLFPGS VIDPPAVNLA AQNKNSSKSR MNPLGSGLAL AISHASHFLQ PPPHQSIIIE RMHSGARRFV TLDFGRPILL TD VLIPTCG DLASLSIDIW TLGEEVDGRR LVVATDISTH SLILHDLIPP PVCRFMKITV IGRYGSTNAR AKIPLGFYYG HSY ILPWES ELKLMHDPLR GEGESASQPE IDQHLAMMVA LQEDIQCRYN LACHRLEALL QSIDLPPLNS ANNAQYFLRK PDKA VEEDS RVFSAYQDCI QLQLQLNLAH NAVQRLKVAI GASRKLLNET SGPEDLIQTS STEQLRTIVR YLLDTLLSLL HSSNG HSVP AVLQSTFHAQ ACEELFKHLC ISGTPKIRLH TGLLLVQLCG GERWWRQFLS NVLQELYNSE QLLIFPQDRV FMLLSC IGQ RSLSNSGVLE SLLNLLDNLL SPLQPELSMH RRTEGVLDIP MISWVVMLVS RLLDYVATVE DEAAAAKKPL NGNQWSF IN NNLHTQNLNR SSKGGSSLDR LYSRKIRKQL VHHKQQLNLL KAKQKALVEQ MEKEKIQSNK GSSYKLLVEQ AKLKQATS K HFKDLIRLRR TAEWSRSNLD TEVTTTKESP EIEPLPFTLA HDRCISVVQK LVLFLLSMDF TCHADLLLFV CKVLARIAN ATRPTIHLCE IVNEPQLERL LLLLVGTDFN RGDISWGGAW AQYSLTCMLQ DILAGELLAP VAAEAMEEGT VSEDVGATAG DSDDSLQQS PAQLLETIDE PLTHEIAGTP PLSSLEKDKE IDLELLQDLM EVDIDPLDID LEKDPLAAKV FKPISSTWYD Y WGADYGTY NYNPYIGGLG MPVAKPPSNT EKNGSQTVSV SVSQALDARL EVGLEQQAEL MLKMMSTLEA DSILQALTNT SP TFSQSPT GTDDSLLGNL QPANQNSQLM IQLSSVPMLN VCFNKLFSML QVHHVQLESL LQLWLTLSLN SSSSGNKENG ADI FLYNAN RIPVISLNQA SIASFLTVLA WYPNTLLRTW CLVLHSLTLM TNMQLNSGSS SSIGIQETTA HLLVSDPNLI HVLV KFLSG TSPHGTNQHS PQVGPTATQA MQEFLTRLQV HLSSTCPQIF SELLLKLIHI LSTERGAFQT GQGPLDAQVK LLEFT LEQN FEVVSVSTIS AVIESVTFLV HHYITCSDKV MSRSGSDSSA GARACFGGLF ANLIRPGDAK AVCGEMTRDQ LMFDLL KLV NILVQLPLSS NREYSARVSV TTNTTDSVSD EEKVSGGKDV NGSSASIPGS PACVADLVLA NQQIMSQILS ALGLCNS SA MAMIIGASGL HLTKHENFHG GLDAISVGDG LFTILTTLSK KASTVHMMLQ PILTYMACGY MGRQGSLATC QLSEPLLW F ILRVLDTSDA LKAFHDMGGV QLICNNMVTS TRAIVNTARS MVSTIMKFLD SGPNKAVDST LKTRILASEP DNAEGIHNF APLGTITSSS PTAQPAEVLL QATPPHRRAR SAAWSYIFLP EEAWCDLTIH LPSAVLLKEI HIQPHLASLA TCPSSVTVEV SADGVNMLP LSTPVVTSGL TYIKIQLVKA EVASAVCLRL HRPRDASTLG LSQIKLLGLT AFGTTSSATV NNPFLPSEDQ V SKTSIGWL RLLHHCLTHI SDLEGMMASA AAPTANLLQT CAALLMSPYC GMHSPNIEVV LVKIGLQSTR IGLKLIDILL RN CAASGSD PTDLNSPLLF GRLNGLSSDS TIDILYQLGT TQDPGTKDRI QALLKWVSDS AKMAALKRSG RMNYMCPSSS AVE YGLLMP SPSHLHCVAA ILWHSYELLV EYDLPALLDR ELFELLFNWS MSLPCNVVLK KAVDSLLCSM CHIHPNYFSL LMGW MGIIP PPVQCHHRLS MTDDSKKQDL SSSLTDDSKN AQAPLSLTES HLATLASSSQ SPEAIKQLLD SGLPSLLVRS LASFC FSHI SYSESIAQSV DNSQDKLRRH HVPQHCNKMP ITADLVAPIL RFLTEVGNSH IMKDWLGGSE VNPLWTALLF LLCHSG STA GGHNLGAQQS STRSASHSSA TTTVLTTQQR TAIENATVAF FLQCISCHPN NQKLMAQVLC ELFQTAPQRG SLPTSGN IS GFVRRLFLQL MLEDEKVTMF LQSPCPLYKG RINATSHVIQ HPMFGAGHKF RTLHLPVSTT LSDVLDRVSD TPSITAKL I SEQKDDKEKK NHEEKEKVKA ENGFQDNYSV VVASGLKSQS KRAVASTPPR PPSRRGRTVP DKIGSASSSA DAASKIITV PVFHLFHRLL AGQPLPAEMT LAQLLTLLYD RKLPQGYRSI DLTVKLGSKV ITDPSLSKTD SFKRLHPEKD HGDLVGSCPE DEALTPSDE CMDGVLDESL LETCPIQSPL QVFAGMGGLA LIAERLPMLY PEVIQQVSAP VIASTTQEKP KDSDQFEWVT I EQSGELVY EAPETIAAEP PPVKSAVQAT SPIPAHSLAA FGLFLRLPGY AEVLLKERKH AQCLLRLVLG VTDDGEGSHI LQ SPSANVL PTLPFHVLRS LFSATPLTTD DGVLLRRMAL EIGALHLILV CLSALSHHAP RVPNSSLSQT EPQVSNSHNP TSA EEQQLY WAKGTGFGTG STASGWDVEQ ALTKQRLEEE HVTCLLQVLA SYINPMSGAV NGEAQASPES RAQNSSALPS MLLE LLSQS CLIPAMSSYL RNDSVLDMAR HVPLYRALLE LLRAIASCTS MVPLLLPLST ENGEEEEDEQ SECQTSVGTL LAKMK TCVD TYTNRLRSKR ENVKAGVKPD APDQEPEGLA LLVPDIQRTA EIVHAATANL RQANQEKKLG EYSKKVVMKP KPLSVL KSL EEKYVAVMKK LQFDTFEMVS EDDDGKLGFK VNYHYMSQVK NANDANSAAR ARRLAQEAVT LSTSLPLSSS SSVFVRC DE ERLDIMKVLI TGPADTPYAN GCFEFDVYFP QDYPSSPPLV NLETTGGHSV RFNPNLYNDG KVCLSILNTW HGRPEEKW N PQTSSFLQVL VSVQSLILVA EPYFNEPGYE RSRGTPSGTQ SSREYDGNIR QATVKWAMLE QIRNPSPCFK EVIHKHFYL KRIELMAQCE EWIADIQQYS SDKRVGRTMS HHAAALKRHT AQLREELLKL PCPEGLDPDI EDASPVCRAT AGAEDTLTHD HVNPSSSKD LPSDFQL UniProtKB: Baculoviral IAP repeat-containing protein 6 |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm Bright-field microscopy / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.0 µm |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 68.4 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 154000 |
 Movie
Movie Controller
Controller











 Z
Z Y
Y X
X