[English] 日本語
 Yorodumi
Yorodumi- EMDB-35740: Cryo-EM structure of SARS-CoV-2 spike protein in complex with dou... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
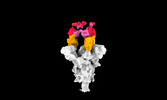
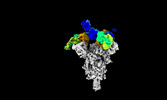
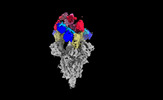
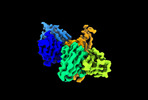
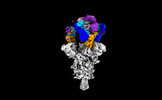
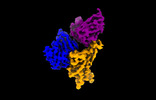
| Title | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Neutralizing antibody / Cryo-EM / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane ...Maturation of spike protein / viral translation / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / suppression by virus of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / entry receptor-mediated virion attachment to host cell / receptor-mediated endocytosis of virus by host cell / Attachment and Entry / membrane fusion / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / receptor ligand activity / host cell surface receptor binding / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.59 Å | |||||||||
 Authors Authors | Sun H / Jiang Y / Zheng Q / Li S / Xia N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2024 Journal: Protein Cell / Year: 2024Title: Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD. Authors: Hui Sun / Tingting Deng / Yali Zhang / Yanling Lin / Yanan Jiang / Yichao Jiang / Yang Huang / Shuo Song / Lingyan Cui / Tingting Li / Hualong Xiong / Miaolin Lan / Liqin Liu / Yu Li / ...Authors: Hui Sun / Tingting Deng / Yali Zhang / Yanling Lin / Yanan Jiang / Yichao Jiang / Yang Huang / Shuo Song / Lingyan Cui / Tingting Li / Hualong Xiong / Miaolin Lan / Liqin Liu / Yu Li / Qianjiao Fang / Kunyu Yu / Wenling Jiang / Lizhi Zhou / Yuqiong Que / Tianying Zhang / Quan Yuan / Tong Cheng / Zheng Zhang / Hai Yu / Jun Zhang / Wenxin Luo / Shaowei Li / Qingbing Zheng / Ying Gu / Ningshao Xia /  Abstract: Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural ...Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural infection or vaccination. The rapid nature of these changes has incited a need for the development of superior broad nAbs (bnAbs) and/or the rational design of an antibody cocktail that can protect against the mutated virus strain. Here, we report two angiotensin-converting enzyme 2 competing nAbs-8H12 and 3E2-with synergistic neutralization but evaded by some Omicron subvariants. Cryo-electron microscopy reveals the two nAbs synergistic neutralizing virus through a rigorous pairing permitted by rearrangement of the 472-489 loop in the receptor-binding domain to avoid steric clashing. Bispecific antibodies based on these two nAbs tremendously extend the neutralizing breadth and restore neutralization against recent variants including currently dominant XBB.1.5. Together, these findings expand our understanding of the potential strategies for the neutralization of SARS-CoV-2 variants toward the design of broad-acting antibody therapeutics and vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35740.map.gz emd_35740.map.gz | 683.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35740-v30.xml emd-35740-v30.xml emd-35740.xml emd-35740.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35740.png emd_35740.png | 41.6 KB | ||
| Filedesc metadata |  emd-35740.cif.gz emd-35740.cif.gz | 6.1 KB | ||
| Others |  emd_35740_half_map_1.map.gz emd_35740_half_map_1.map.gz emd_35740_half_map_2.map.gz emd_35740_half_map_2.map.gz | 675.5 MB 675.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35740 http://ftp.pdbj.org/pub/emdb/structures/EMD-35740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35740 | HTTPS FTP |
-Validation report
| Summary document |  emd_35740_validation.pdf.gz emd_35740_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35740_full_validation.pdf.gz emd_35740_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35740_validation.xml.gz emd_35740_validation.xml.gz | 20.4 KB | Display | |
| Data in CIF |  emd_35740_validation.cif.gz emd_35740_validation.cif.gz | 24.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35740 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35740 | HTTPS FTP |
-Related structure data
| Related structure data |  8iv4MC  8iv5C  8iv8C  8ivaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35740.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35740.map.gz / Format: CCP4 / Size: 729 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.778 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35740_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
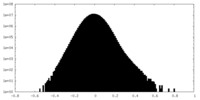
| Projections & Slices |
| ||||||||||||
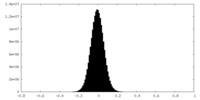
| Density Histograms |
-Half map: #2
| File | emd_35740_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
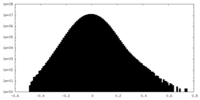
| Projections & Slices |
| ||||||||||||
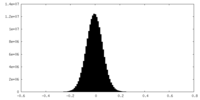
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 spike protein in complex with dou...
| Entire | Name: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 spike protein in complex with dou...
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: SARS-CoV-2 spike protein
| Supramolecule | Name: SARS-CoV-2 spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: nAbs 8H12 and 3E2
| Supramolecule | Name: nAbs 8H12 and 3E2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: light chain of 3E2
| Macromolecule | Name: light chain of 3E2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.203357 KDa |
| Sequence | String: QIVLTQSPAI MSASLGEEIT LTCSVSSSVS DMHWYQQKSG TSPKVFIYST SNLASGVPSR FSGSGSGTFY SLTISSVEAE DAAYYYCHQ WSSWTFGGGT KLEIK |
-Macromolecule #2: heavy chain of 3E2
| Macromolecule | Name: heavy chain of 3E2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.105676 KDa |
| Sequence | String: EVMLVESGGG VVKPGGSLKL SCAASGFSFS TYAMSWIRQT PEKSLEWVAA ISSGGTNTYY PGSVKGRFTI SRDKAMNTLY LQLSSLRSE DTAMYYCVRH SGNYVDSVMD YWGQGTSVTV SS |
-Macromolecule #3: light chain of 8H12
| Macromolecule | Name: light chain of 8H12 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.945558 KDa |
| Sequence | String: DIVMTQFQKF MSTSVGDRVS ITCKASQNVR TAVAWYQQKP GQSPKAMIYL ASNRHRGVPD RFTGSGCGTD FTLTISNVQC EDLADYFCL QHRNYPLTFG GGTKLEIK |
-Macromolecule #4: heavy chain of 8H12
| Macromolecule | Name: heavy chain of 8H12 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.240784 KDa |
| Sequence | String: EVKLEESGGG LVQPGGSMKL SCAASGFTFS DAWMDWVRQS PEKGLEWVAQ IRRKANNHAT YYAESVKGRF TISRDDSKSS VYLQMNSLR AEDTGIYYCI RGMTYAMDFW GQGTSVTVSS |
-Macromolecule #5: Spike protein S1
| Macromolecule | Name: Spike protein S1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.930721 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ESIVRFPNIT NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAP GQTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG V EGFNCYFP ...String: ESIVRFPNIT NLCPFGEVFN ATRFASVYAW NRKRISNCVA DYSVLYNSAS FSTFKCYGVS PTKLNDLCFT NVYADSFVIR GDEVRQIAP GQTGKIADYN YKLPDDFTGC VIAWNSNNLD SKVGGNYNYL YRLFRKSNLK PFERDISTEI YQAGSTPCNG V EGFNCYFP LQSYGFQPTN GVGYQPYRVV VLSFELLHAP ATVCGP UniProtKB: Spike glycoprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.59 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 217835 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller















 Z
Z Y
Y X
X