[English] 日本語
 Yorodumi
Yorodumi- EMDB-35731: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SARS-CoV-2 / Neutralizing antibody / Cryo-EM / VIRAL PROTEIN | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.57 Å | |||||||||
 Authors Authors | Sun H / Jiang Y / Zheng Q / Li S / Xia N | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2024 Journal: Protein Cell / Year: 2024Title: Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD. Authors: Hui Sun / Tingting Deng / Yali Zhang / Yanling Lin / Yanan Jiang / Yichao Jiang / Yang Huang / Shuo Song / Lingyan Cui / Tingting Li / Hualong Xiong / Miaolin Lan / Liqin Liu / Yu Li / ...Authors: Hui Sun / Tingting Deng / Yali Zhang / Yanling Lin / Yanan Jiang / Yichao Jiang / Yang Huang / Shuo Song / Lingyan Cui / Tingting Li / Hualong Xiong / Miaolin Lan / Liqin Liu / Yu Li / Qianjiao Fang / Kunyu Yu / Wenling Jiang / Lizhi Zhou / Yuqiong Que / Tianying Zhang / Quan Yuan / Tong Cheng / Zheng Zhang / Hai Yu / Jun Zhang / Wenxin Luo / Shaowei Li / Qingbing Zheng / Ying Gu / Ningshao Xia /  Abstract: Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural ...Continual evolution of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) virus has allowed for its gradual evasion of neutralizing antibodies (nAbs) produced in response to natural infection or vaccination. The rapid nature of these changes has incited a need for the development of superior broad nAbs (bnAbs) and/or the rational design of an antibody cocktail that can protect against the mutated virus strain. Here, we report two angiotensin-converting enzyme 2 competing nAbs-8H12 and 3E2-with synergistic neutralization but evaded by some Omicron subvariants. Cryo-electron microscopy reveals the two nAbs synergistic neutralizing virus through a rigorous pairing permitted by rearrangement of the 472-489 loop in the receptor-binding domain to avoid steric clashing. Bispecific antibodies based on these two nAbs tremendously extend the neutralizing breadth and restore neutralization against recent variants including currently dominant XBB.1.5. Together, these findings expand our understanding of the potential strategies for the neutralization of SARS-CoV-2 variants toward the design of broad-acting antibody therapeutics and vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35731.map.gz emd_35731.map.gz | 119.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35731-v30.xml emd-35731-v30.xml emd-35731.xml emd-35731.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35731.png emd_35731.png | 19 KB | ||
| Filedesc metadata |  emd-35731.cif.gz emd-35731.cif.gz | 4.1 KB | ||
| Others |  emd_35731_half_map_1.map.gz emd_35731_half_map_1.map.gz emd_35731_half_map_2.map.gz emd_35731_half_map_2.map.gz | 226.7 MB 226.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35731 http://ftp.pdbj.org/pub/emdb/structures/EMD-35731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35731 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35731 | HTTPS FTP |
-Validation report
| Summary document |  emd_35731_validation.pdf.gz emd_35731_validation.pdf.gz | 762.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35731_full_validation.pdf.gz emd_35731_full_validation.pdf.gz | 762.3 KB | Display | |
| Data in XML |  emd_35731_validation.xml.gz emd_35731_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_35731_validation.cif.gz emd_35731_validation.cif.gz | 19 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35731 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35731 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35731 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35731 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35731.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35731.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.12 Å | ||||||||||||||||||||||||||||||||||||
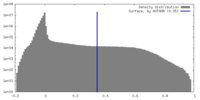
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_35731_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
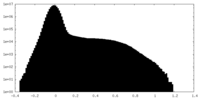
| Projections & Slices |
| ||||||||||||
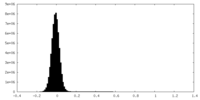
| Density Histograms |
-Half map: #2
| File | emd_35731_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
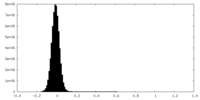
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2
| Entire | Name: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2 |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2
| Supramolecule | Name: Cryo-EM structure of SARS-CoV-2 spike protein in complex with 3E2 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|
-Supramolecule #2: SARS-CoV-2 spike protein
| Supramolecule | Name: SARS-CoV-2 spike protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: nAb 3E2
| Supramolecule | Name: nAb 3E2 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#7 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.57 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 17367 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)