[English] 日本語
 Yorodumi
Yorodumi- EMDB-35364: Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/R... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
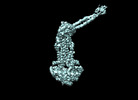
| Title | Cryo-EM structure of Mycobacterium tuberculosis ATP bound FtsEX/RipC complex in peptidisc | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | complex / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on peptide bonds (peptidases) / transmembrane transporter activity / cysteine-type peptidase activity / transmembrane transport / manganese ion binding / cell division / magnesium ion binding / ATP hydrolysis activity / proteolysis / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
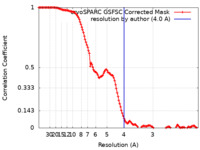
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Li J / Xu X / Luo M | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Regulation of the cell division hydrolase RipC by the FtsEX system in Mycobacterium tuberculosis. Authors: Jianwei Li / Xin Xu / Jian Shi / Juan A Hermoso / Lok-To Sham / Min Luo /   Abstract: The FtsEX complex regulates, directly or via a protein mediator depending on bacterial genera, peptidoglycan degradation for cell division. In mycobacteria and Gram-positive bacteria, the FtsEX ...The FtsEX complex regulates, directly or via a protein mediator depending on bacterial genera, peptidoglycan degradation for cell division. In mycobacteria and Gram-positive bacteria, the FtsEX system directly activates peptidoglycan-hydrolases by a mechanism that remains unclear. Here we report our investigation of Mycobacterium tuberculosis FtsEX as a non-canonical regulator with high basal ATPase activity. The cryo-EM structures of the FtsEX system alone and in complex with RipC, as well as the ATP-activated state, unveil detailed information on the signal transduction mechanism, leading to the activation of RipC. Our findings indicate that RipC is recognized through a "Match and Fit" mechanism, resulting in an asymmetric rearrangement of the extracellular domains of FtsX and a unique inclined binding mode of RipC. This study provides insights into the molecular mechanisms of FtsEX and RipC regulation in the context of a critical human pathogen, guiding the design of drugs targeting peptidoglycan remodeling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35364.map.gz emd_35364.map.gz | 173.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35364-v30.xml emd-35364-v30.xml emd-35364.xml emd-35364.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35364_fsc.xml emd_35364_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_35364.png emd_35364.png | 37.1 KB | ||
| Filedesc metadata |  emd-35364.cif.gz emd-35364.cif.gz | 5.9 KB | ||
| Others |  emd_35364_half_map_1.map.gz emd_35364_half_map_1.map.gz emd_35364_half_map_2.map.gz emd_35364_half_map_2.map.gz | 169.7 MB 169.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35364 http://ftp.pdbj.org/pub/emdb/structures/EMD-35364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35364 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35364 | HTTPS FTP |
-Validation report
| Summary document |  emd_35364_validation.pdf.gz emd_35364_validation.pdf.gz | 984.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35364_full_validation.pdf.gz emd_35364_full_validation.pdf.gz | 984.5 KB | Display | |
| Data in XML |  emd_35364_validation.xml.gz emd_35364_validation.xml.gz | 21.6 KB | Display | |
| Data in CIF |  emd_35364_validation.cif.gz emd_35364_validation.cif.gz | 28.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35364 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35364 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35364 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35364 | HTTPS FTP |
-Related structure data
| Related structure data |  8iddMC  8idbC  8idcC  8igqC  8jiaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35364.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35364.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35364_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
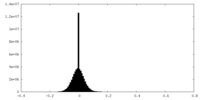
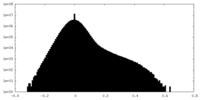
| Density Histograms |
-Half map: #1
| File | emd_35364_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
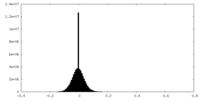
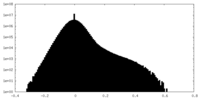
| Density Histograms |
- Sample components
Sample components
-Entire : complex of FtsEX-RipC-ATP
| Entire | Name: complex of FtsEX-RipC-ATP |
|---|---|
| Components |
|
-Supramolecule #1: complex of FtsEX-RipC-ATP
| Supramolecule | Name: complex of FtsEX-RipC-ATP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cell division ATP-binding protein FtsE
| Macromolecule | Name: Cell division ATP-binding protein FtsE / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.766697 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MMITLDHVTK QYKSSARPAL DDINVKIDKG EFVFLIGPSG SGKSTFMRLL LAAETPTSGD VRVSKFHVNK LRGRHVPKLR QVIGCVFQD FRLLQQKTVY DNVAFALEVI GKRTDAINRV VPEVLETVGL SGKANRLPDE LSGGEQQRVA IARAFVNRPL V LLADEPTG ...String: MMITLDHVTK QYKSSARPAL DDINVKIDKG EFVFLIGPSG SGKSTFMRLL LAAETPTSGD VRVSKFHVNK LRGRHVPKLR QVIGCVFQD FRLLQQKTVY DNVAFALEVI GKRTDAINRV VPEVLETVGL SGKANRLPDE LSGGEQQRVA IARAFVNRPL V LLADEPTG NLDPETSRDI MDLLERINRT GTTVLMATHD HHIVDSMRQR VVELSLGRLV RDEQRGVYGM DR UniProtKB: Cell division ATP-binding protein FtsE |
-Macromolecule #2: Cell division protein FtsX
| Macromolecule | Name: Cell division protein FtsX / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.851355 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRFGFLLNEV LTGFRRNVTM TIAMILTTAI SVGLFGGGML VVRLADSSRA IYLDRVESQV FLTEDVSAND SSCDTTACKA LREKIETRS DVKAVRFLNR QQAYDDAIRK FPQFKDVAGK DSFPASFIVK LENPEQHKDF DTAMKGQPGV LDVLNQKELI D RLFAVLDG ...String: MRFGFLLNEV LTGFRRNVTM TIAMILTTAI SVGLFGGGML VVRLADSSRA IYLDRVESQV FLTEDVSAND SSCDTTACKA LREKIETRS DVKAVRFLNR QQAYDDAIRK FPQFKDVAGK DSFPASFIVK LENPEQHKDF DTAMKGQPGV LDVLNQKELI D RLFAVLDG LSNAAFAVAL VQAIGAILLI ANMVQVAAYT RRTEIGIMRL VGASRWYTQL PFLVEAMLAA TMGVGIAVAG LM VVRALFL ENALNQFYQA NLIAKVDYAD ILFITPWLLL LGVAMSGLTA YLTLRLYVRR UniProtKB: Cell division protein FtsX |
-Macromolecule #3: Probable endopeptidase MT2245
| Macromolecule | Name: Probable endopeptidase MT2245 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.792918 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRLDQRWLIA RVIMRSAIGF FASFTVSSGV LAANVLADPA DDALAKLNEL SRQAEQTTEA LHSAQLDLNE KLAAQRAADQ KLADNRTAL DAARARLATF QTAVNKVAAA TYMGGRTHGM DAILTAESPQ LLIDRLSVQR VMAHQMSTQM ARFKAAGEQA V KAEQAAAK ...String: MRLDQRWLIA RVIMRSAIGF FASFTVSSGV LAANVLADPA DDALAKLNEL SRQAEQTTEA LHSAQLDLNE KLAAQRAADQ KLADNRTAL DAARARLATF QTAVNKVAAA TYMGGRTHGM DAILTAESPQ LLIDRLSVQR VMAHQMSTQM ARFKAAGEQA V KAEQAAAK SAADARSAAE QAAAVRANLQ HKQSQLQVQI AVVKSQYVAL TPEERTALAD PGPVPAVAAI APGAPPAALP PG APPGDGP APGVAPPPGG MPGLPFVQPD GAGGDRTAVV QAALTQVGAP YAWGGAAPGG FDCSGLVMWA FQQAGIALPH SSQ ALAHGG QPVALSDLQP GDVLTFYSDA SHAGIYIGDG LMVHSSTYGV PVRVVPMDSS GPIYDARRY UniProtKB: Probable endopeptidase MT2245 |
-Macromolecule #4: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 6.02 sec. / Average electron dose: 38.837 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)