+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Lb2Cas12a RNA DNA complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Lachnospiraceae bacterium MA2020 (bacteria) / synthetic construct (others) Lachnospiraceae bacterium MA2020 (bacteria) / synthetic construct (others) | |||||||||
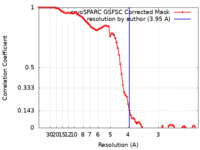
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 3.95 Å cryo EM / Resolution: 3.95 Å | |||||||||
 Authors Authors | Li J / Sivaraman J / Satoru M | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2023 Journal: PLoS Biol / Year: 2023Title: Structures of apo Cas12a and its complex with crRNA and DNA reveal the dynamics of ternary complex formation and target DNA cleavage. Authors: Li Jianwei / Chacko Jobichen / Satoru Machida / Sun Meng / Randy J Read / Chen Hongying / Shi Jian / Yuren Adam Yuan / J Sivaraman /   Abstract: Cas12a is a programmable nuclease for adaptive immunity against invading nucleic acids in CRISPR-Cas systems. Here, we report the crystal structures of apo Cas12a from Lachnospiraceae bacterium ...Cas12a is a programmable nuclease for adaptive immunity against invading nucleic acids in CRISPR-Cas systems. Here, we report the crystal structures of apo Cas12a from Lachnospiraceae bacterium MA2020 (Lb2) and the Lb2Cas12a+crRNA complex, as well as the cryo-EM structure and functional studies of the Lb2Cas12a+crRNA+DNA complex. We demonstrate that apo Lb2Cas12a assumes a unique, elongated conformation, whereas the Lb2Cas12a+crRNA binary complex exhibits a compact conformation that subsequently rearranges to a semi-open conformation in the Lb2Cas12a+crRNA+DNA ternary complex. Notably, in solution, apo Lb2Cas12a is dynamic and can exist in both elongated and compact forms. Residues from Met493 to Leu523 of the WED domain undergo major conformational changes to facilitate the required structural rearrangements. The REC lobe of Lb2Cas12a rotates 103° concomitant with rearrangement of the hinge region close to the WED and RuvC II domains to position the RNA-DNA duplex near the catalytic site. Our findings provide insight into crRNA recognition and the mechanism of target DNA cleavage. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35191.map.gz emd_35191.map.gz | 31.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35191-v30.xml emd-35191-v30.xml emd-35191.xml emd-35191.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35191_fsc.xml emd_35191_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_35191.png emd_35191.png | 60.1 KB | ||
| Others |  emd_35191_additional_1.map.gz emd_35191_additional_1.map.gz emd_35191_half_map_1.map.gz emd_35191_half_map_1.map.gz emd_35191_half_map_2.map.gz emd_35191_half_map_2.map.gz | 2.1 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35191 http://ftp.pdbj.org/pub/emdb/structures/EMD-35191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35191 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35191.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35191.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.105 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #1
| File | emd_35191_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
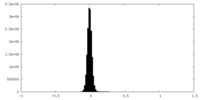
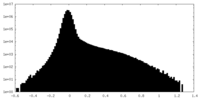
| Density Histograms |
-Half map: #2
| File | emd_35191_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
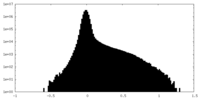
| Density Histograms |
-Half map: #1
| File | emd_35191_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
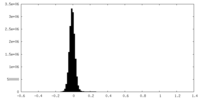
| Density Histograms |
- Sample components
Sample components
-Entire : LbCas12a-crRNA-DNA ternary complex
| Entire | Name: LbCas12a-crRNA-DNA ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: LbCas12a-crRNA-DNA ternary complex
| Supramolecule | Name: LbCas12a-crRNA-DNA ternary complex / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium MA2020 (bacteria) Lachnospiraceae bacterium MA2020 (bacteria) |
| Molecular weight | Theoretical: 140 KDa |
-Macromolecule #1: Lb2Cas12a
| Macromolecule | Name: Lb2Cas12a / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium MA2020 (bacteria) Lachnospiraceae bacterium MA2020 (bacteria) |
| Molecular weight | Theoretical: 141.200609 KDa |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Sequence | String: MYYESLTKQY PVSKTIRNEL IPIGKTLDNI RQNNILESDV KRKQNYEHVK GILDEYHKQL INEALDNCTL PSLKIAAEIY LKNQKEVSD REDFNKTQDL LRKEVVEKLK AHENFTKIGK KDILDLLEKL PSISEDDYNA LESFRNFYTY FTSYNKVREN L YSDKEKSS ...String: MYYESLTKQY PVSKTIRNEL IPIGKTLDNI RQNNILESDV KRKQNYEHVK GILDEYHKQL INEALDNCTL PSLKIAAEIY LKNQKEVSD REDFNKTQDL LRKEVVEKLK AHENFTKIGK KDILDLLEKL PSISEDDYNA LESFRNFYTY FTSYNKVREN L YSDKEKSS TVAYRLINEN FPKFLDNVKS YRFVKTAGIL ADGLGEEEQD SLFIVETFNK TLTQDGIDTY NSQVGKINSS IN LYNQKNQ KANGFRKIPK MKMLYKQILS DREESFIDEF QSDEVLIDNV ESYGSVLIES LKSSKVSAFF DALRESKGKN VYV KNDLAK TAMSNIVFEN WRTFDDLLNQ EYDLANENKK KDDKYFEKRQ KELKKNKSYS LEHLCNLSED SCNLIENYIH QISD DIENI IINNETFLRI VINEHDRSRK LAKNRKAVKA IKDFLDSIKV LERELKLINS SGQELEKDLI VYSAHEELLV ELKQV DSLY NMTRNYLTKK PFSTEKVKLN FNRSTLLNGW DRNKETDNLG VLLLKDGKYY LGIMNTSANK AFVNPPVAKT EKVFKK VDY KLLPVPNQML PKVFFAKSNI DFYNPSSEIY SNYKKGTHKK GNMFSLEDCH NLIDFFKESI SKHEDWSKFG FKFSDTA SY NDISEFYREV EKQGYKLTYT DIDETYINDL IERNELYLFQ IYNKDFSMYS KGKLNLHTLY FMMLFDQRNI DDVVYKLN G EAEVFYRPAS ISEDELIIHK AGEEIKNKNP NRARTKETST FSYDIVKDKR YSKDKFTLHI PITMNFGVDE VKRFNDAVN SAIRIDENVN VIGIDRGERN LLYVVVIDSK GNILEQISLN SIINKEYDIE TDYHALLDER EGGRDKARKD WNTVENIRDL KAGYLSQVV NVVAKLVLKY NAIICLEDLN FGFKRGRQKV EKQVYQKFEK MLIDKLNYLV IDKSREQTSP KELGGALNAL Q LTSKFKSF KELGKQSGVI YYVPAYLTSK IDPTTGFANL FYMKCENVEK SKRFFDGFDF IRFNALENVF EFGFDYRSFT QR ACGINSK WTVCTNGERI IKYRNPDKNN MFDEKVVVVT DEMKNLFEQY KIPYEDGRNV KDMIISNEEA EFYRRLYRLL QQT LQMRNS TSDGTRDYII SPVKNKREAY FNSELSDGSV PKDADANGAY NIARKGLWVL EQIRQKSEGE KINLAMTNAE WLEY AQTHL L |
-Macromolecule #2: RNA (33-MER)
| Macromolecule | Name: RNA (33-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Lachnospiraceae bacterium MA2020 (bacteria) Lachnospiraceae bacterium MA2020 (bacteria) |
| Molecular weight | Theoretical: 10.452219 KDa |
| Sequence | String: AAUUUCUACU AAUUGUAGAU GCCGCUACCC CGA |
-Macromolecule #3: DNA (25-MER)
| Macromolecule | Name: DNA (25-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 7.780007 KDa |
| Sequence | String: (DT)(DG)(DG)(DT)(DC)(DG)(DG)(DG)(DG)(DT) (DA)(DG)(DC)(DG)(DG)(DC)(DT)(DA)(DA)(DA) (DG)(DC)(DA)(DC)(DT) |
-Macromolecule #4: DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3')
| Macromolecule | Name: DNA (5'-D(*AP*GP*TP*GP*CP*TP*TP*TP*A)-3') / type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 2.745821 KDa |
| Sequence | String: (DA)(DG)(DT)(DG)(DC)(DT)(DT)(DT)(DA) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 50 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295.15 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Temperature | Min: 140.0 K / Max: 150.0 K |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 2560 / Average exposure time: 3.49 sec. / Average electron dose: 45.0 e/Å2 |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X