+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CnCas12f1-sgRNA-DNA complex | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Complex / CRISPR-Cas / RNA BINDING PROTEIN/RNA/DNA / RNA BINDING PROTEIN-RNA-DNA complex | ||||||||||||
| Function / homology | Transposase IS605, OrfB, C-terminal / Putative transposase DNA-binding domain / transposition / DNA recombination / DNA binding / Transposase Function and homology information Function and homology information | ||||||||||||
| Biological species |  Clostridium novyi (bacteria) / synthetic construct (others) Clostridium novyi (bacteria) / synthetic construct (others) | ||||||||||||
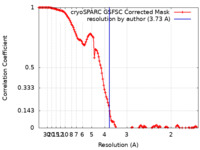
| Method | single particle reconstruction / Resolution: 3.73 Å | ||||||||||||
 Authors Authors | Li F / Ji Q | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure and engineering of miniature Clostridium novyi CRISPR-Cas12f1 with rare C-rich PAM specificity Authors: Li F / Ji Q | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34958.map.gz emd_34958.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34958-v30.xml emd-34958-v30.xml emd-34958.xml emd-34958.xml | 15.4 KB 15.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34958_fsc.xml emd_34958_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_34958.png emd_34958.png | 111.3 KB | ||
| Others |  emd_34958_half_map_1.map.gz emd_34958_half_map_1.map.gz emd_34958_half_map_2.map.gz emd_34958_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34958 http://ftp.pdbj.org/pub/emdb/structures/EMD-34958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34958 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34958 | HTTPS FTP |
-Validation report
| Summary document |  emd_34958_validation.pdf.gz emd_34958_validation.pdf.gz | 821.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34958_full_validation.pdf.gz emd_34958_full_validation.pdf.gz | 820.9 KB | Display | |
| Data in XML |  emd_34958_validation.xml.gz emd_34958_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_34958_validation.cif.gz emd_34958_validation.cif.gz | 28 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34958 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34958 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34958 | HTTPS FTP |
-Related structure data
| Related structure data |  8hr5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34958.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34958.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34958_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34958_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of CnCas12f1-sgRNA-DNA
| Entire | Name: Ternary complex of CnCas12f1-sgRNA-DNA |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of CnCas12f1-sgRNA-DNA
| Supramolecule | Name: Ternary complex of CnCas12f1-sgRNA-DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Clostridium novyi (bacteria) Clostridium novyi (bacteria) |
-Macromolecule #1: Transposase
| Macromolecule | Name: Transposase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Clostridium novyi (bacteria) Clostridium novyi (bacteria) |
| Molecular weight | Theoretical: 58.60968 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MITVRKIKLT IMGDKDTRNS QYKWIRDEQY NQYRALNMGM TYLAVNDILY MNESGLEIRT IKDLKDCEKD IDKNKKEIEK LTARLEKEQ NKKNSSSEKL DEIKYKISLV ENKIEDYKLK IVELNKILEE TQKERMDIQK EFKEKYVDDL YQVLDKIPFK H LDNKSLVT ...String: MITVRKIKLT IMGDKDTRNS QYKWIRDEQY NQYRALNMGM TYLAVNDILY MNESGLEIRT IKDLKDCEKD IDKNKKEIEK LTARLEKEQ NKKNSSSEKL DEIKYKISLV ENKIEDYKLK IVELNKILEE TQKERMDIQK EFKEKYVDDL YQVLDKIPFK H LDNKSLVT QRIKADIKSD KSNGLLKGER SIRNYKRNFP LMTRGRDLKF KYDDNDDIEI KWMEGIKFKV ILGNRIKNSL EL RHTLHKV IEGKYKICDS SLQFDKNNNL ILNLTLDIPI DIVNKKVSGR VVGVDLGLKI PAYCALNDVE YIKKSIGRID DFL KVRTQM QSRRRRLQIA IQSAKGGKGR VNKLQALERF AEKEKNFAKT YNHFLSSNIV KFAVSNQAEQ INMELLSLKE TQNK SILRN WSYYQLQTMI EYKAQREGIK VKYIDPYHTS QTCSKCGNYE EGQRESQADF ICKKCGYKVN ADYNAARNIA MSNKY ITKK EESKYYKIKE SMV UniProtKB: Transposase |
-Macromolecule #2: sgRNA
| Macromolecule | Name: sgRNA / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 75.498406 KDa |
| Sequence | String: AACAUAGUUA AACUAAUAAA AACAGGGCGA UUUAACGUCC UAAGGCUGAG AGAAGUUUUU UCUACUCGGC AAGGGUUAAU CUCGAUUGU UGUGUUACCG AUCGAGCGUU UCACAAAAUG CGAGAGAAAU CUCGCAUUUU UAAUUUUGCA GUAAGGCUAG U UUUUAUAU ...String: AACAUAGUUA AACUAAUAAA AACAGGGCGA UUUAACGUCC UAAGGCUGAG AGAAGUUUUU UCUACUCGGC AAGGGUUAAU CUCGAUUGU UGUGUUACCG AUCGAGCGUU UCACAAAAUG CGAGAGAAAU CUCGCAUUUU UAAUUUUGCA GUAAGGCUAG U UUUUAUAU AAAUAUGCUA UAACUAGGGU UUUAGUUUAA CUAUGUGAAA UGUAAAUAAU AGAUGUGAAG UCGCUUU |
-Macromolecule #3: DNA (28-MER)
| Macromolecule | Name: DNA (28-MER) / type: dna / ID: 3 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 8.612591 KDa |
| Sequence | String: (DG)(DA)(DA)(DA)(DA)(DG)(DC)(DG)(DA)(DC) (DT)(DT)(DC)(DA)(DC)(DA)(DT)(DC)(DT)(DA) (DT)(DT)(DA)(DG)(DG)(DT)(DT)(DA) |
-Macromolecule #4: DNA (5'-D(*TP*AP*AP*CP*CP*TP*AP*AP*TP*AP*GP*AP*TP*GP*TP*GP*AP*A)-3')
| Macromolecule | Name: DNA (5'-D(*TP*AP*AP*CP*CP*TP*AP*AP*TP*AP*GP*AP*TP*GP*TP*GP*AP*A)-3') type: dna / ID: 4 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 5.547643 KDa |
| Sequence | String: (DT)(DA)(DA)(DC)(DC)(DT)(DA)(DA)(DT)(DA) (DG)(DA)(DT)(DG)(DT)(DG)(DA)(DA) |
-Experimental details
-Structure determination
 Processing Processing | single particle reconstruction |
|---|---|
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)